Plot Box-and-Whisker plots (Tukey 1970; McGill et al. 1978) to graphically compare the probability distributions of quantitative traits between entire collection (EC) and core set (CS).
Arguments
- data
The data as a data frame object. The data frame should possess one row per individual and columns with the individual names and multiple trait/character data.
- names
Name of column with the individual names as a character string.
- quantitative
Name of columns with the quantitative traits as a character vector.
- selected
Character vector with the names of individuals selected in core collection and present in the
namescolumn.- show.count
logical. If
TRUE, the accession count excluding missing values will also be displayed. Default isFALSE.
Value
A list with the ggplot objects of box plots of CS and EC for
each trait specified as quantitative.
References
McGill R, Tukey JW, Larsen WA (1978).
“Variations of box plots.”
The American Statistician, 32(1), 12.
Tukey JW (1970).
Exploratory Data Analysis. Preliminary edition.
Addison-Wesley.
Examples
data("cassava_CC")
data("cassava_EC")
ec <- cbind(genotypes = rownames(cassava_EC), cassava_EC)
ec$genotypes <- as.character(ec$genotypes)
rownames(ec) <- NULL
core <- rownames(cassava_CC)
quant <- c("NMSR", "TTRN", "TFWSR", "TTRW", "TFWSS", "TTSW", "TTPW", "AVPW",
"ARSR", "SRDM")
qual <- c("CUAL", "LNGS", "PTLC", "DSTA", "LFRT", "LBTEF", "CBTR", "NMLB",
"ANGB", "CUAL9M", "LVC9M", "TNPR9M", "PL9M", "STRP", "STRC",
"PSTR")
ec[, qual] <- lapply(ec[, qual],
function(x) factor(as.factor(x)))
# \donttest{
box.evaluate.core(data = ec, names = "genotypes",
quantitative = quant, selected = core)
#> $NMSR
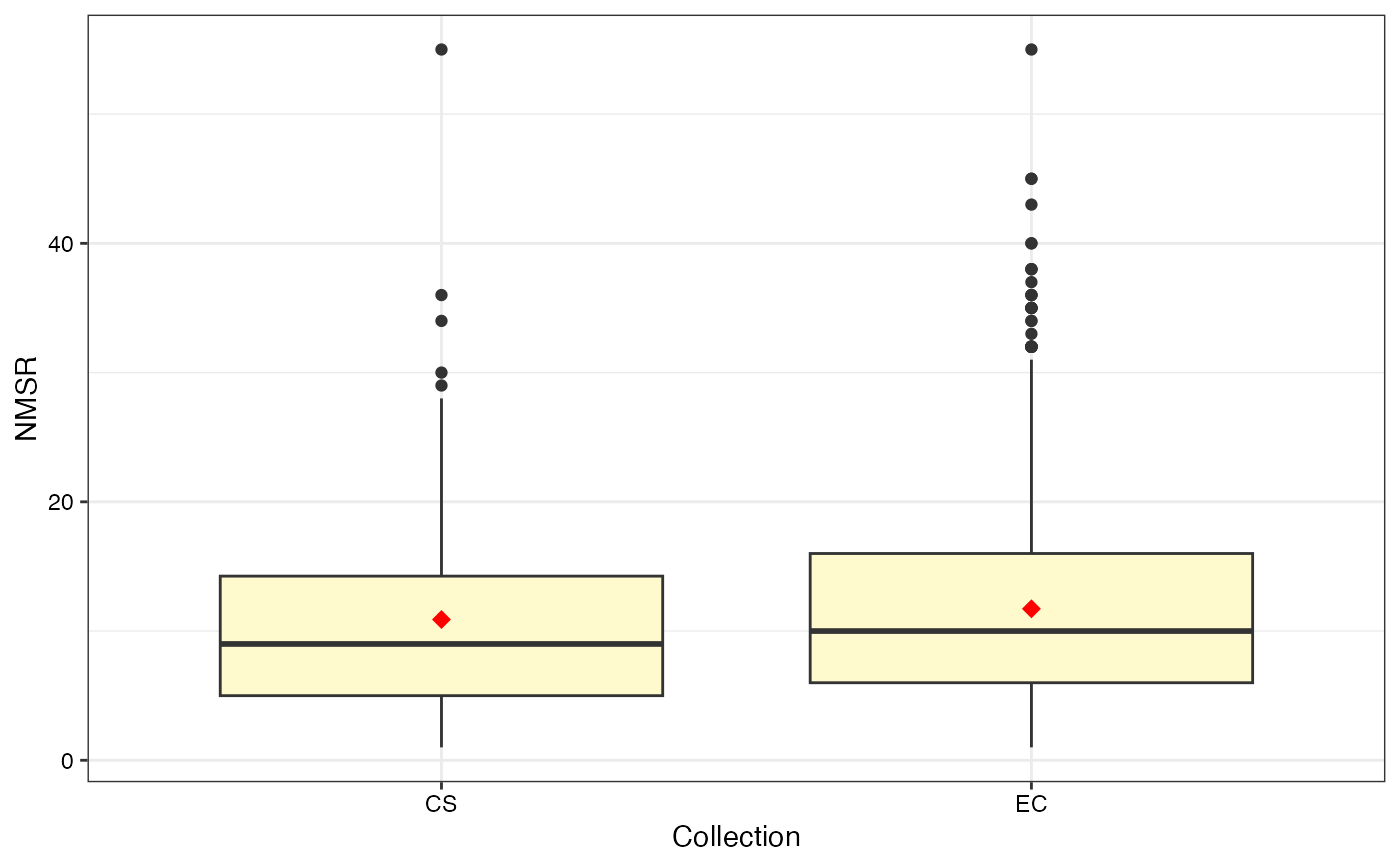 #>
#> $TTRN
#>
#> $TTRN
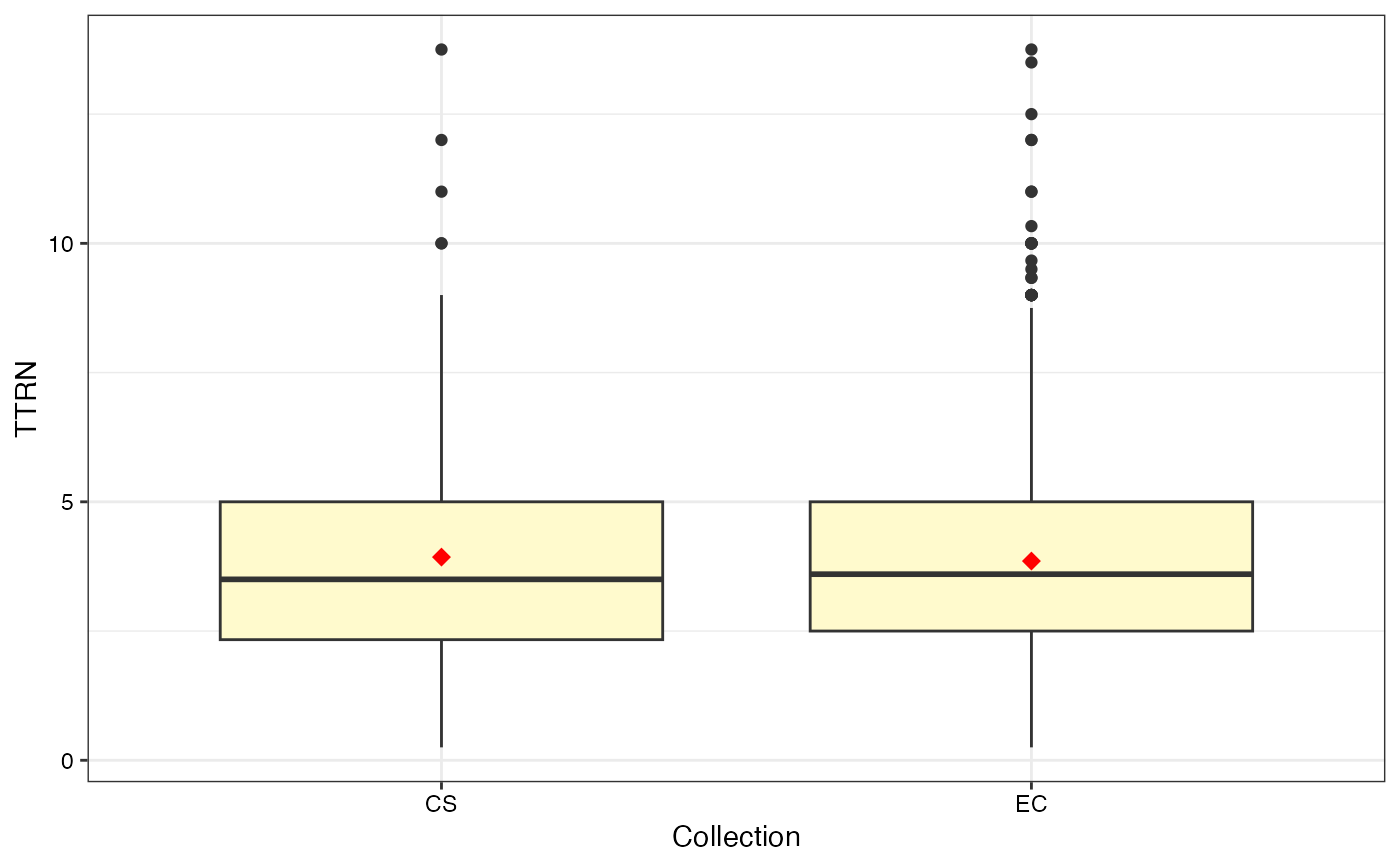 #>
#> $TFWSR
#>
#> $TFWSR
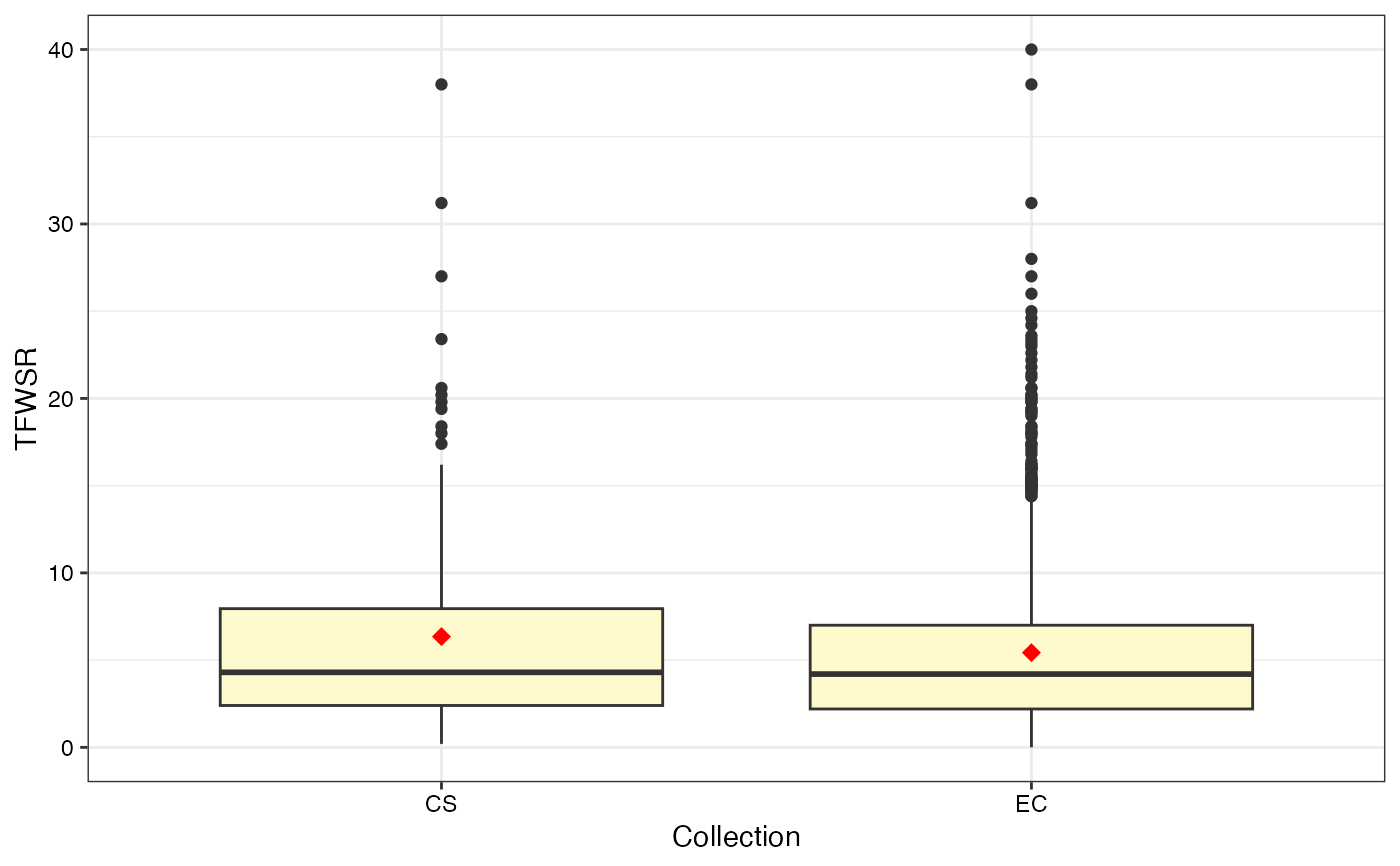 #>
#> $TTRW
#>
#> $TTRW
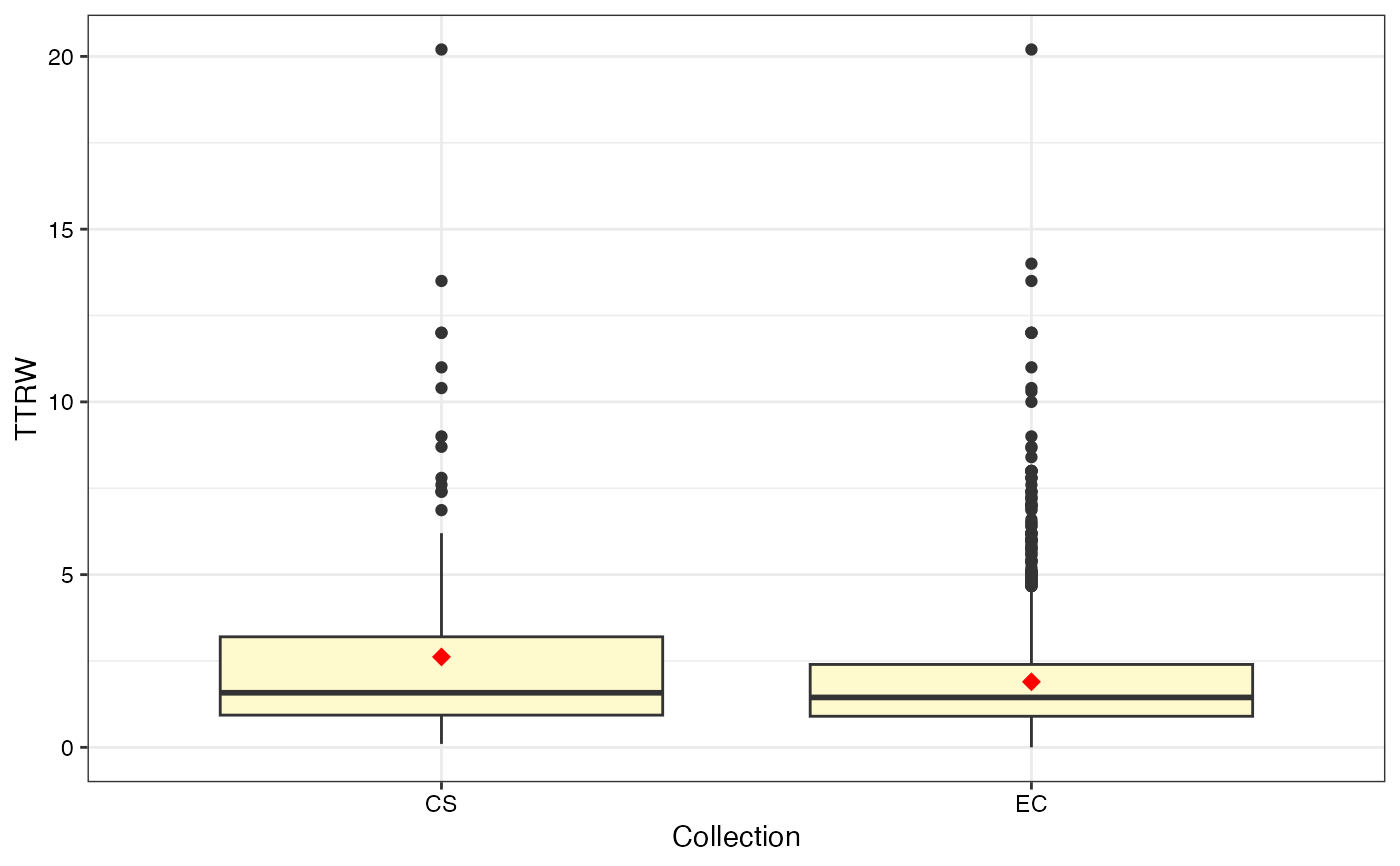 #>
#> $TFWSS
#>
#> $TFWSS
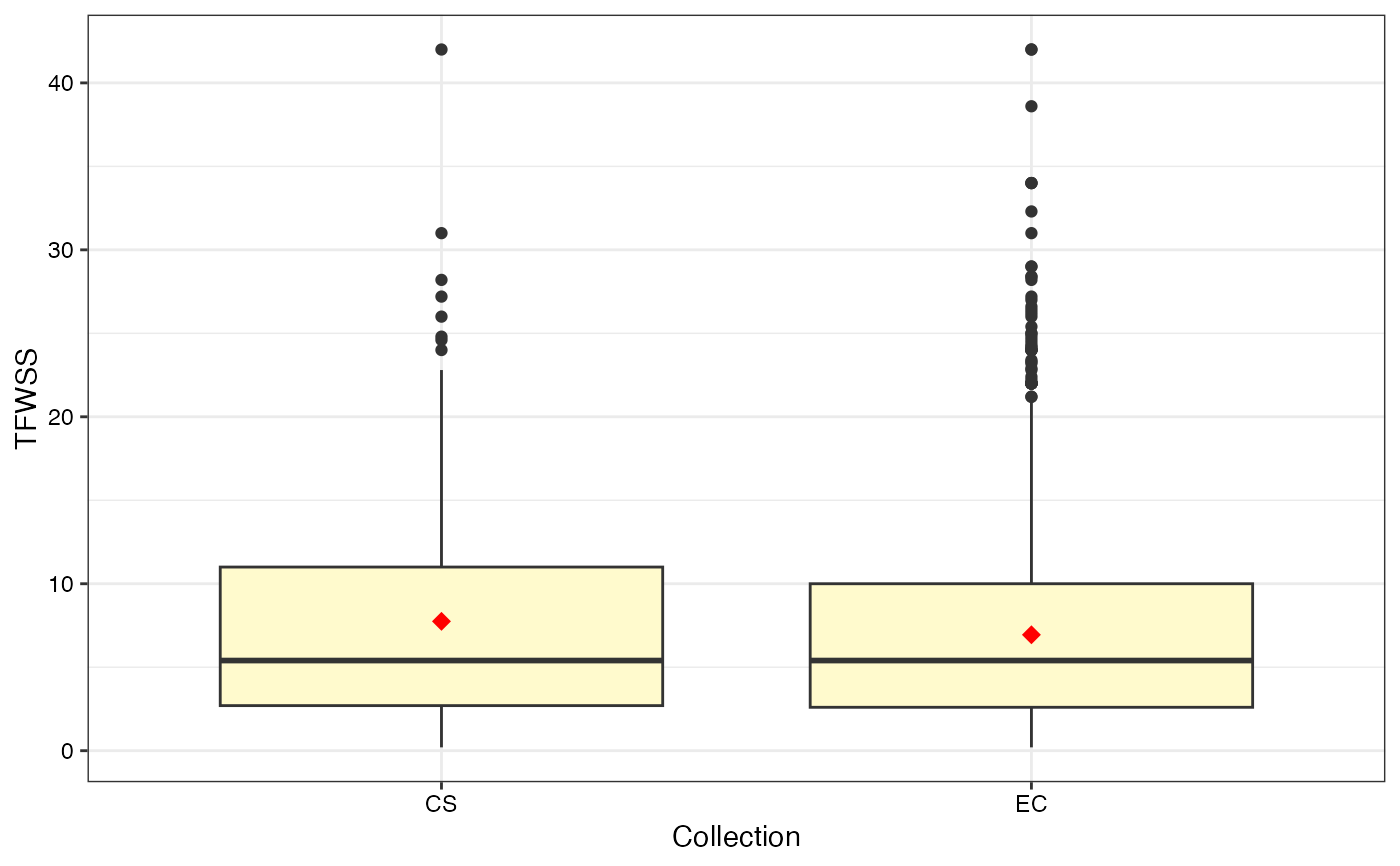 #>
#> $TTSW
#>
#> $TTSW
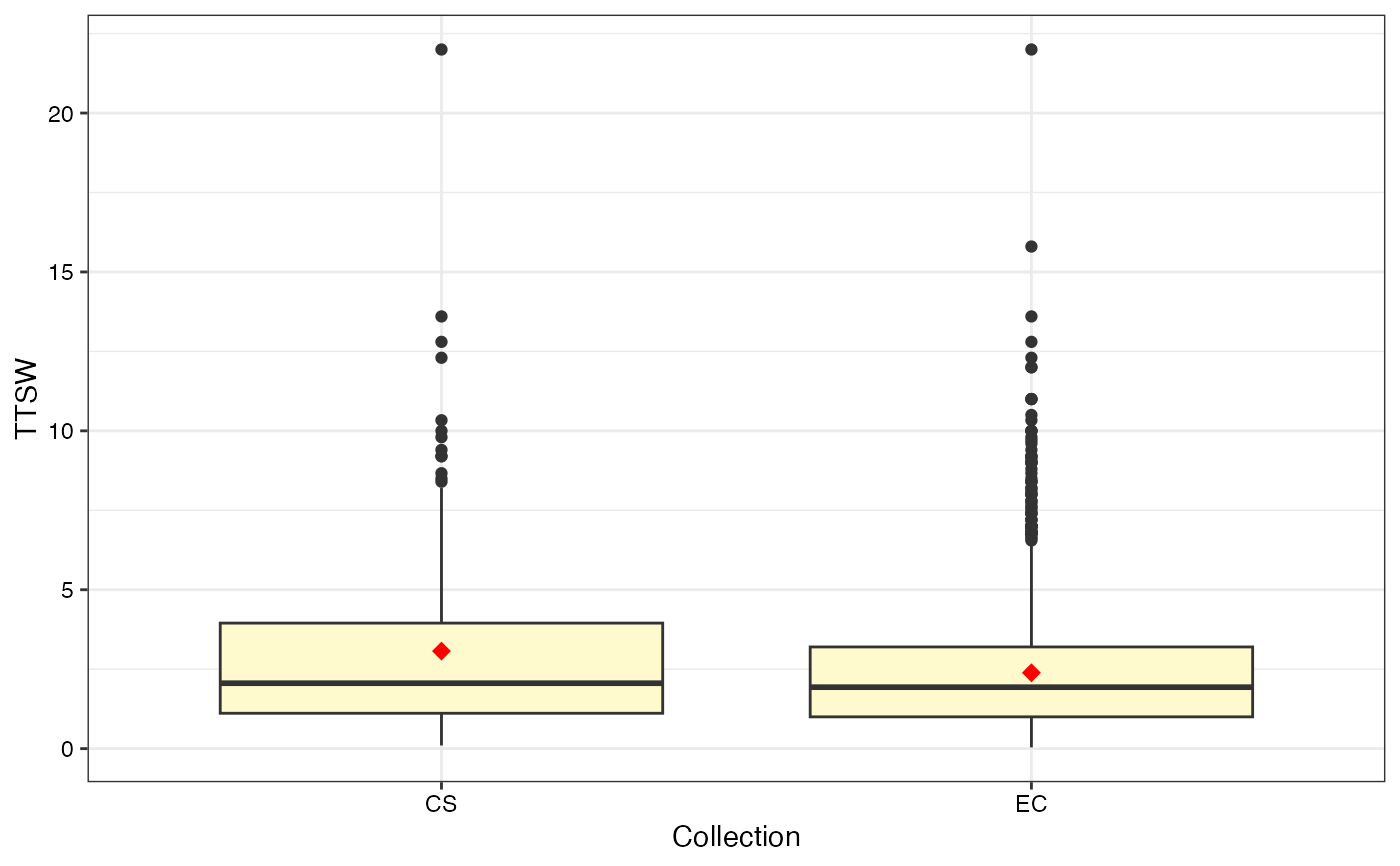 #>
#> $TTPW
#>
#> $TTPW
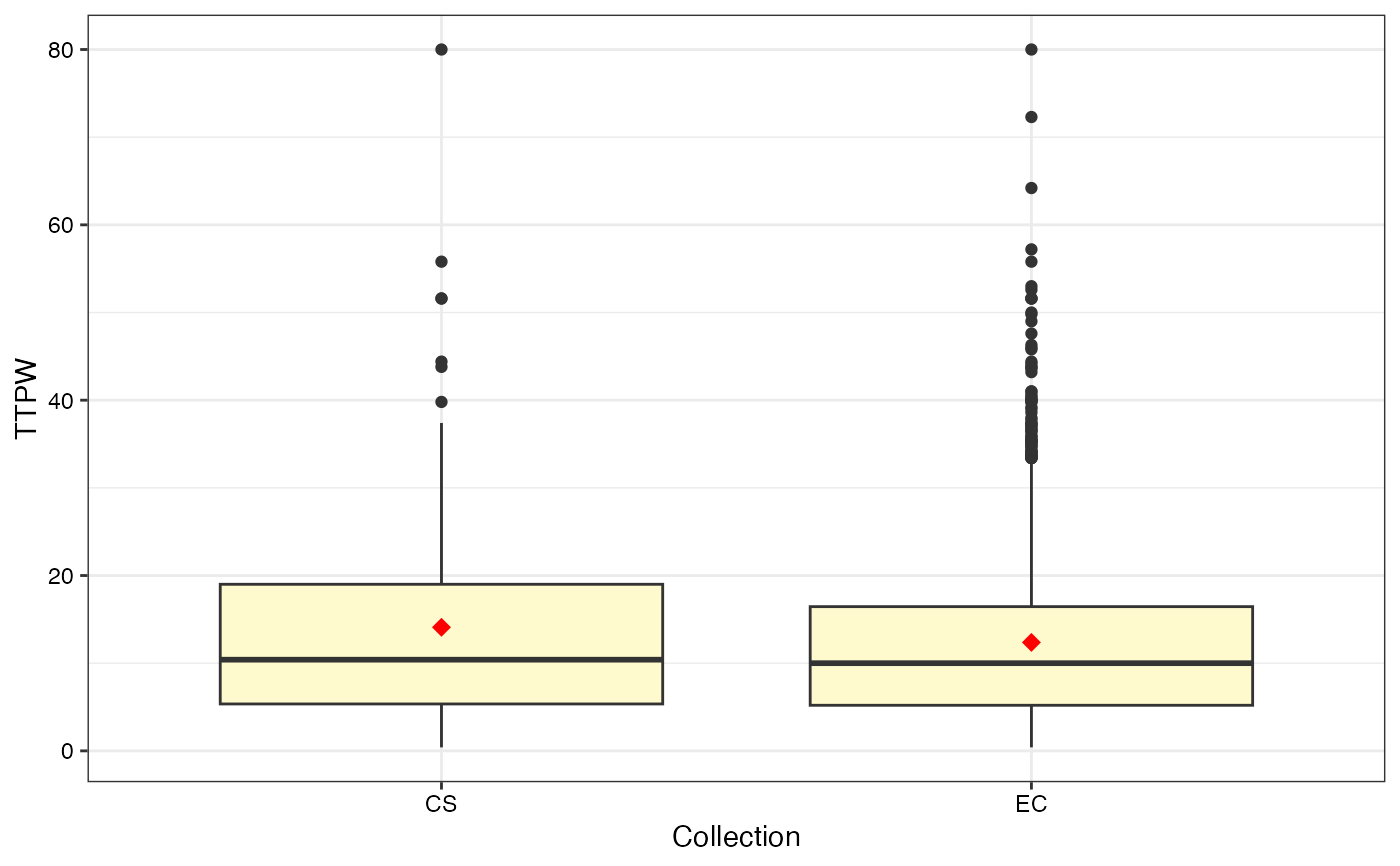 #>
#> $AVPW
#>
#> $AVPW
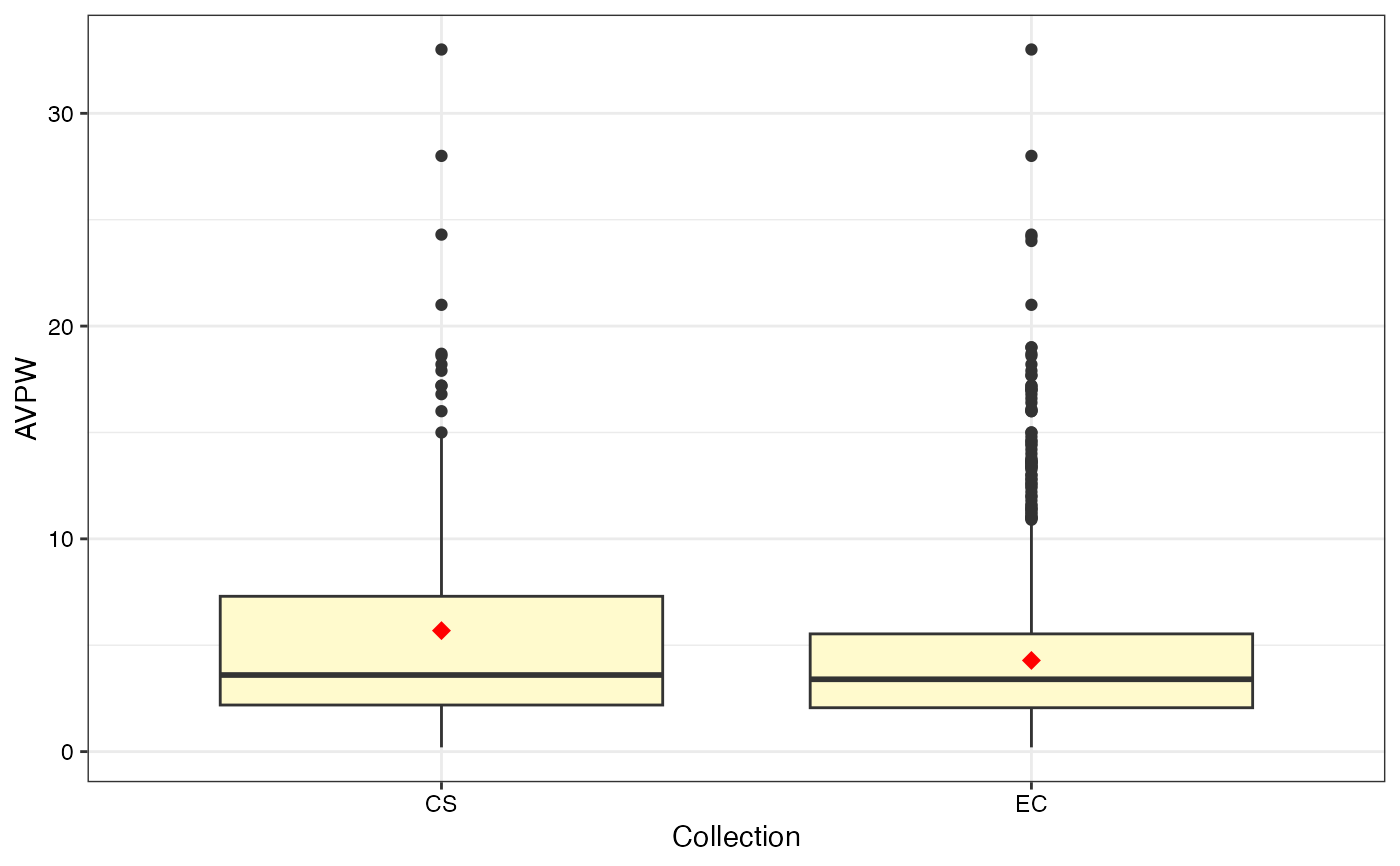 #>
#> $ARSR
#>
#> $ARSR
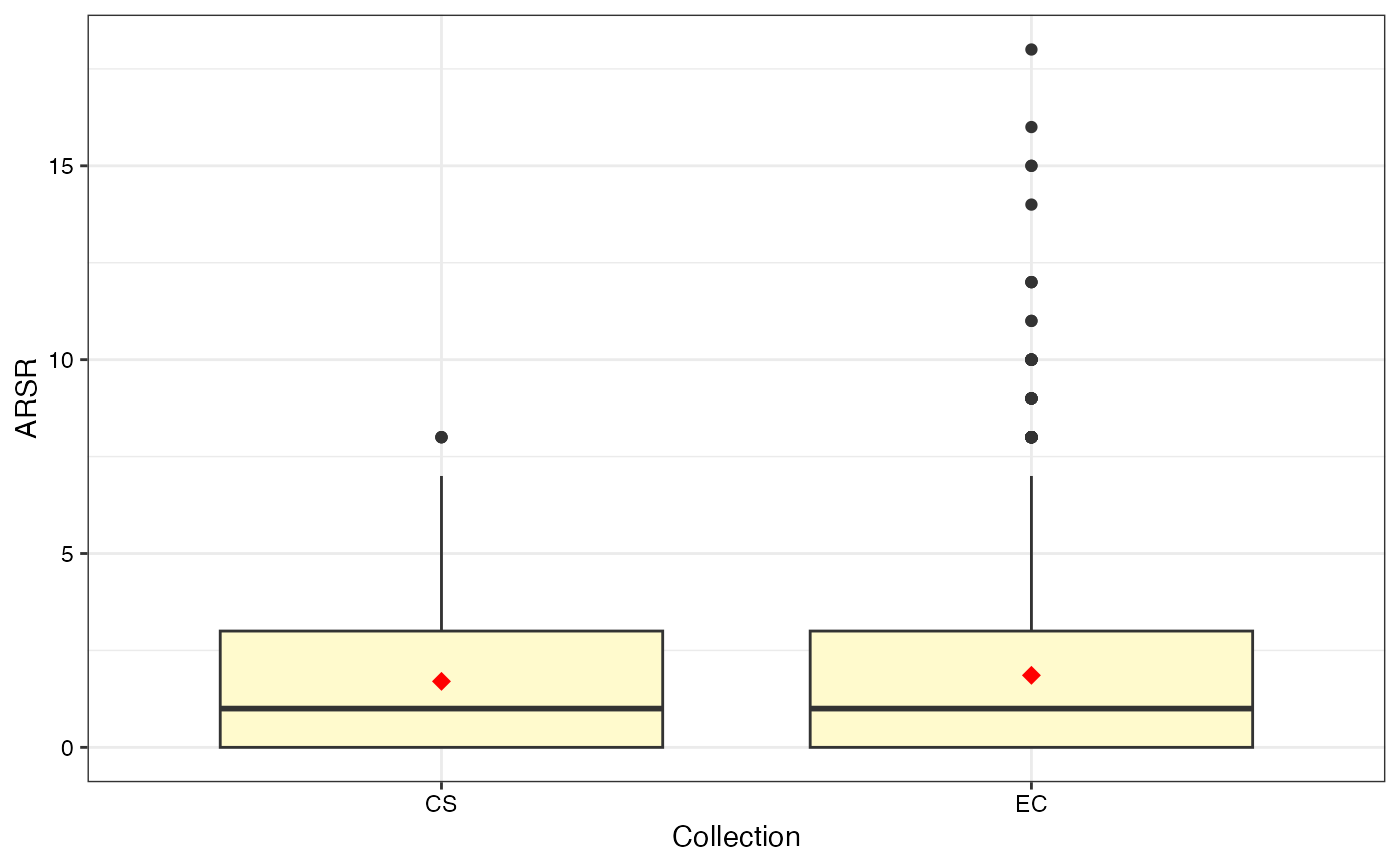 #>
#> $SRDM
#>
#> $SRDM
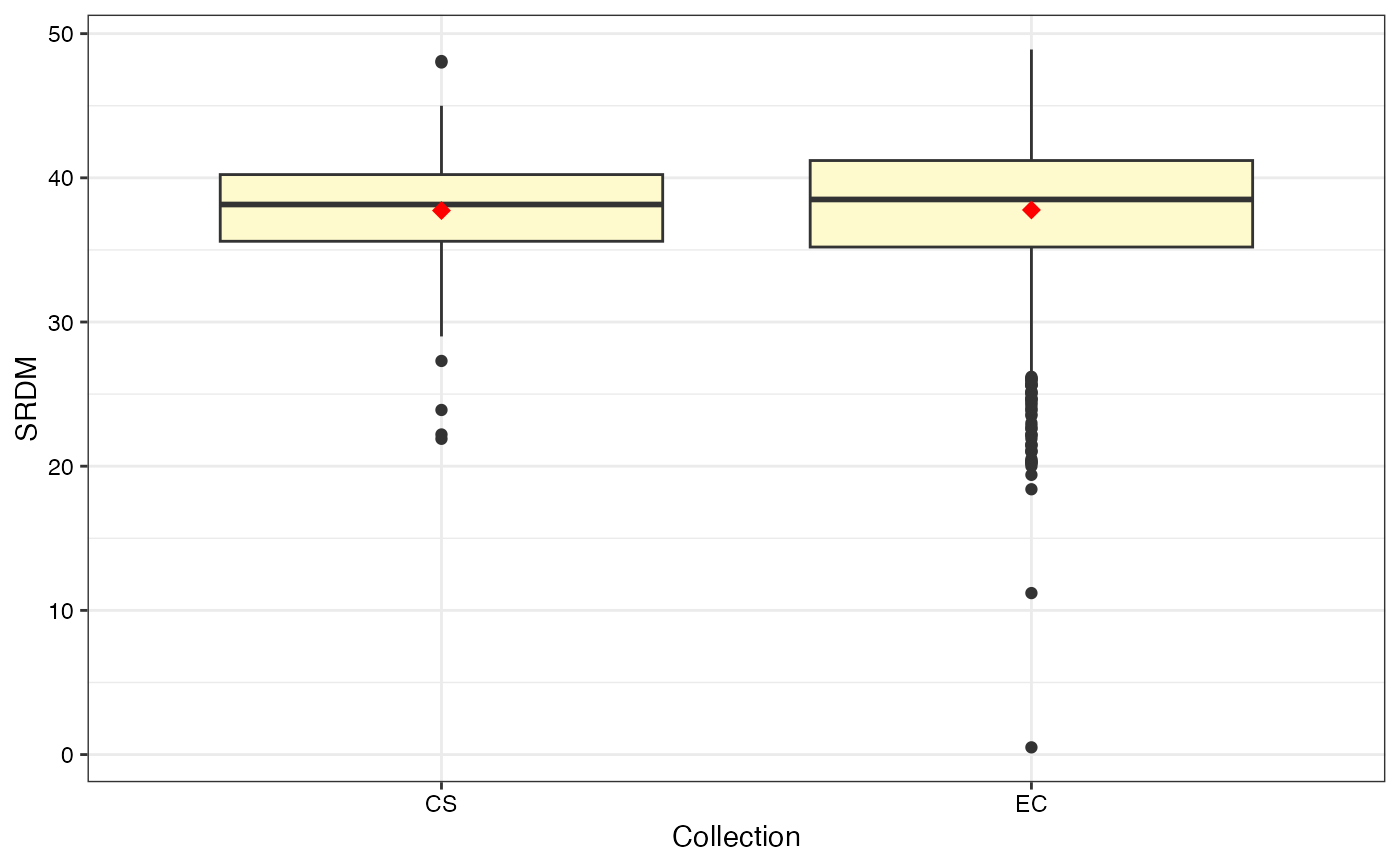 #>
box.evaluate.core(data = ec, names = "genotypes",
quantitative = quant, selected = core, show.count = TRUE)
#> $NMSR
#>
box.evaluate.core(data = ec, names = "genotypes",
quantitative = quant, selected = core, show.count = TRUE)
#> $NMSR
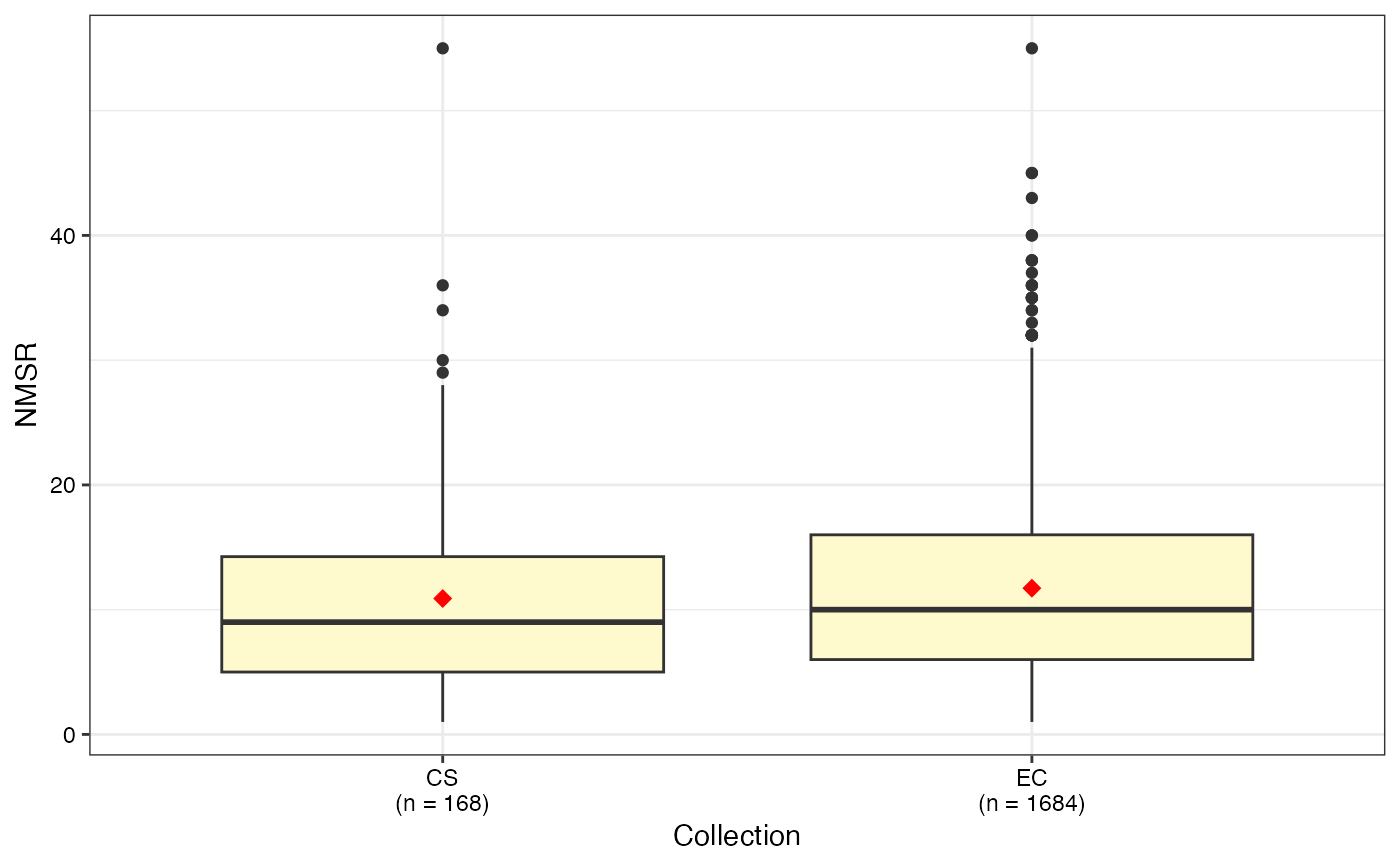 #>
#> $TTRN
#>
#> $TTRN
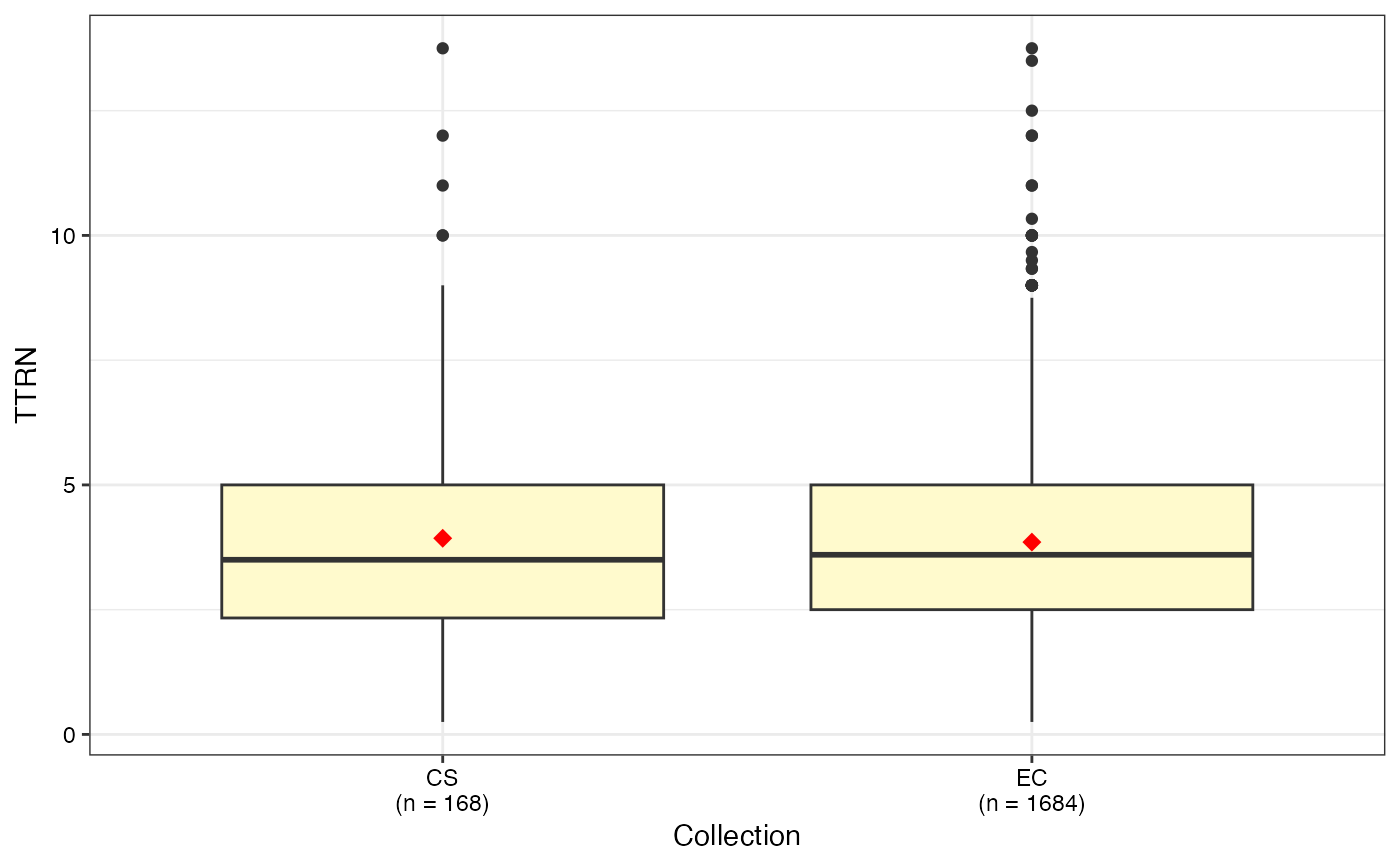 #>
#> $TFWSR
#>
#> $TFWSR
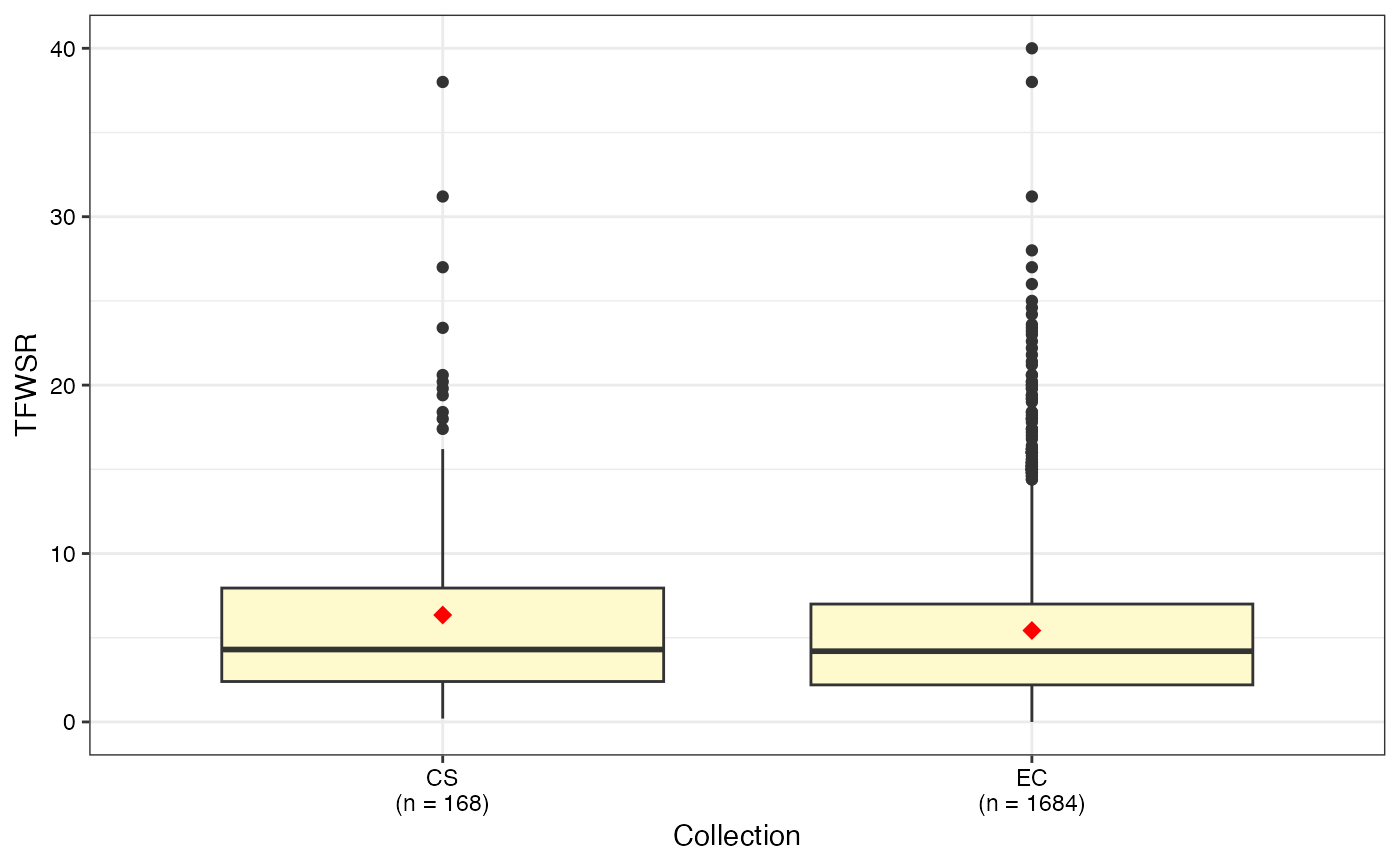 #>
#> $TTRW
#>
#> $TTRW
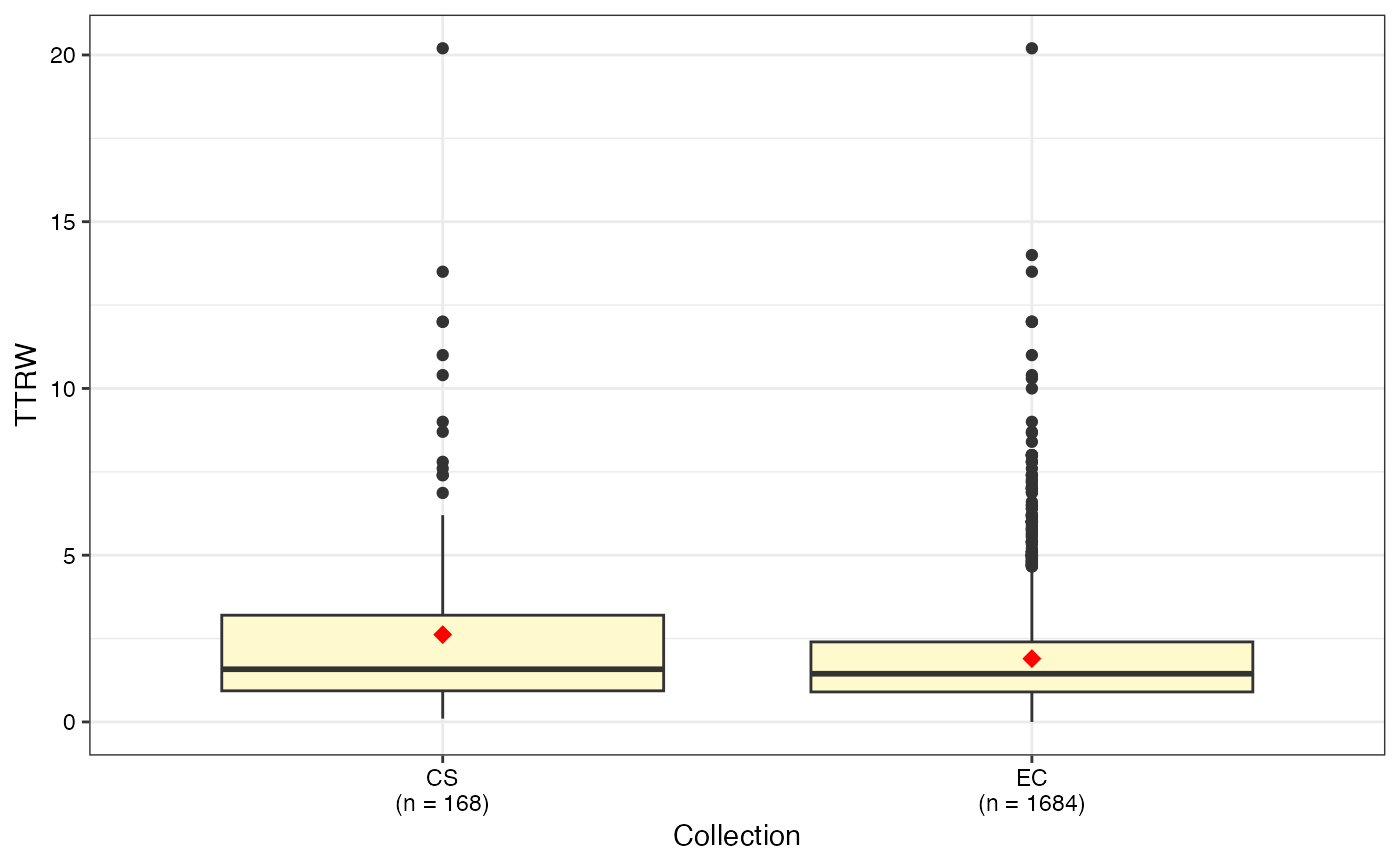 #>
#> $TFWSS
#>
#> $TFWSS
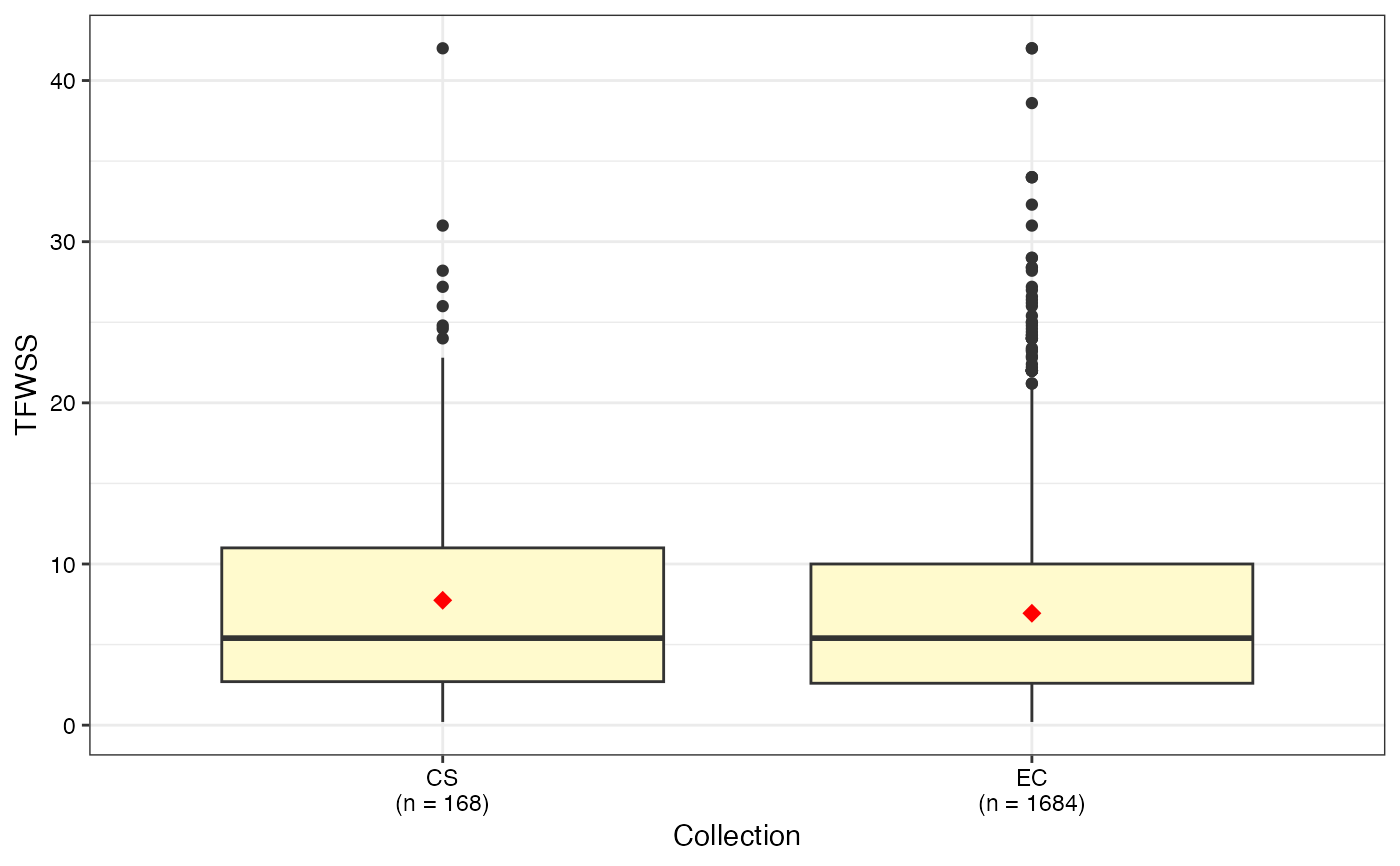 #>
#> $TTSW
#>
#> $TTSW
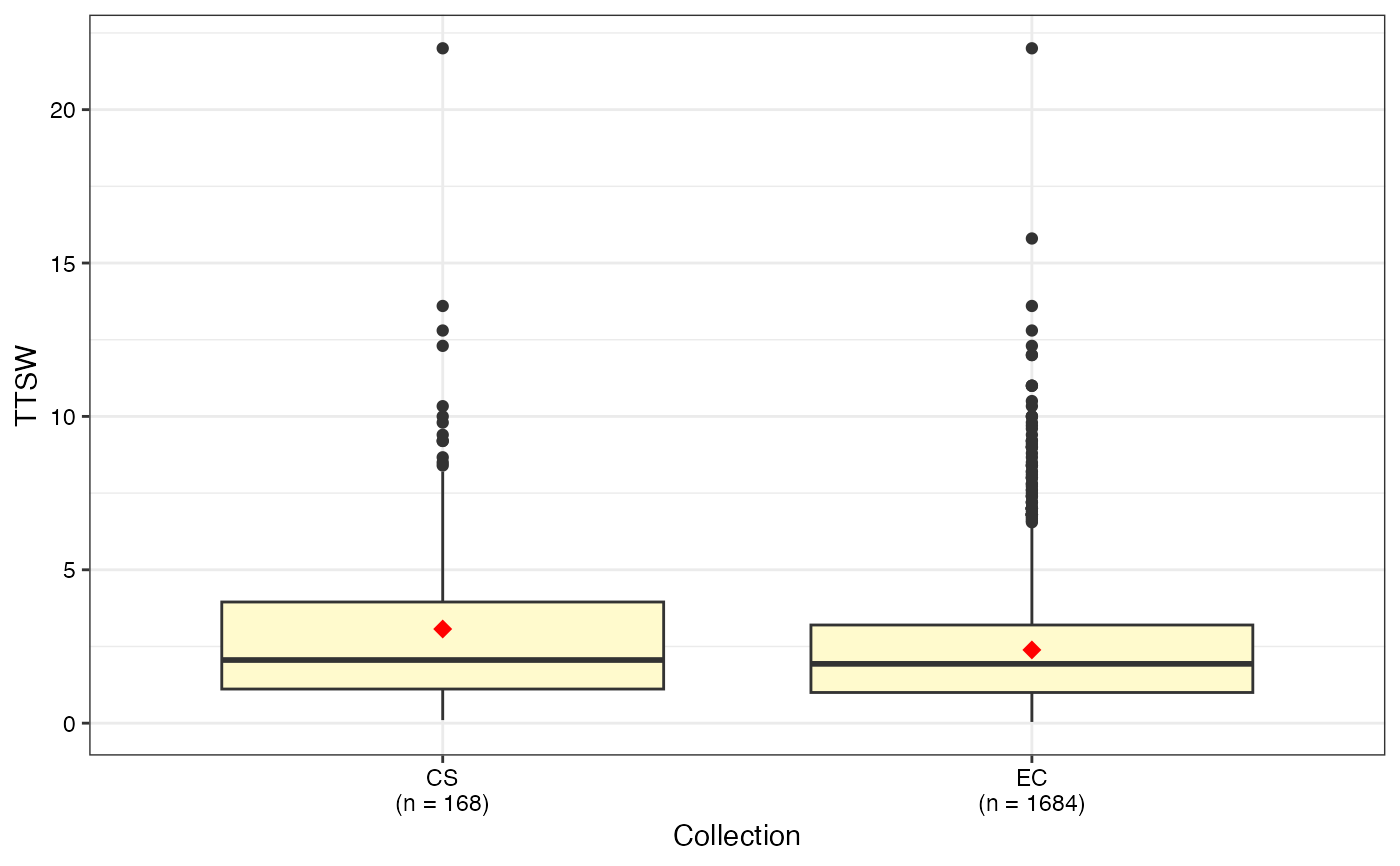 #>
#> $TTPW
#>
#> $TTPW
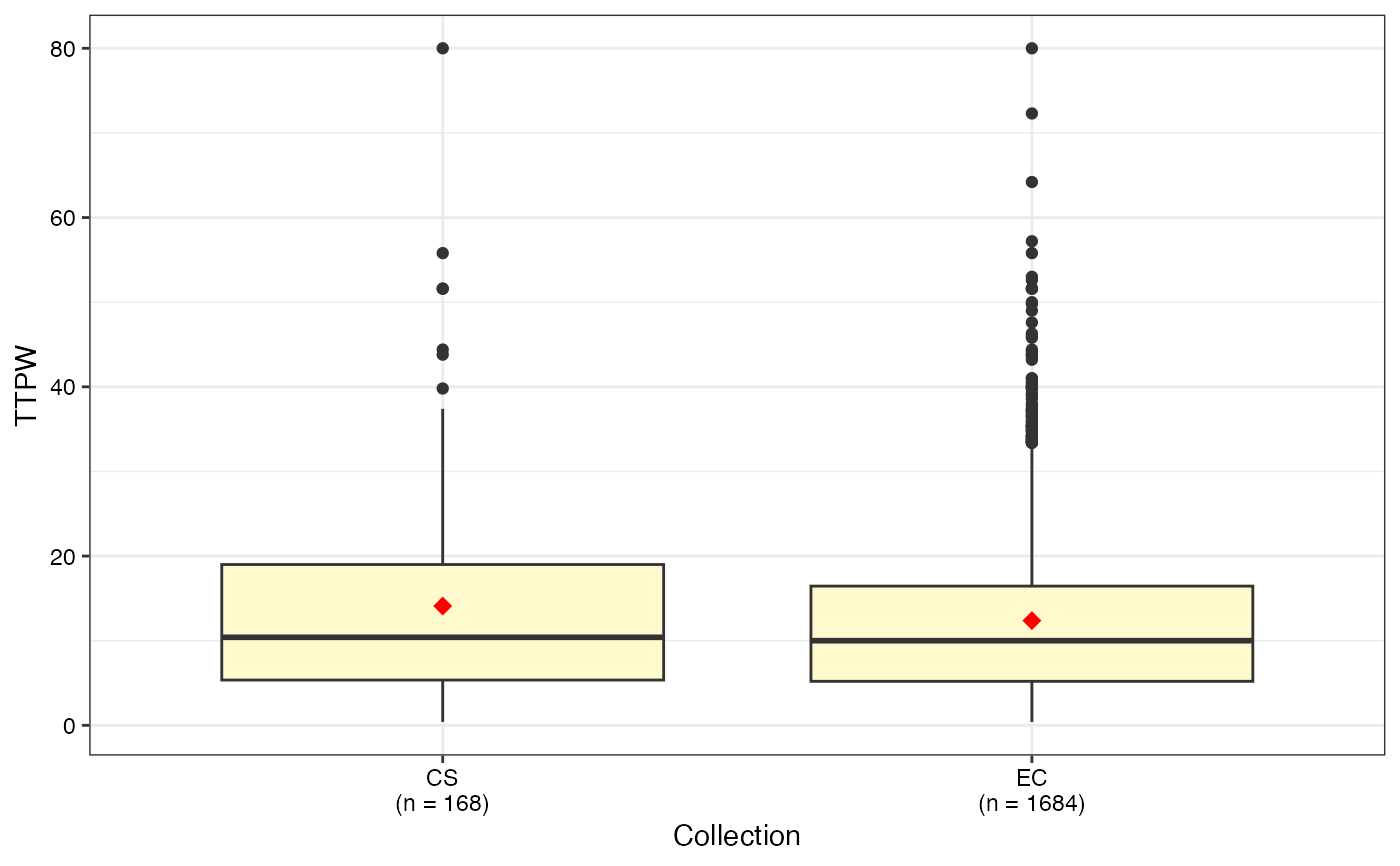 #>
#> $AVPW
#>
#> $AVPW
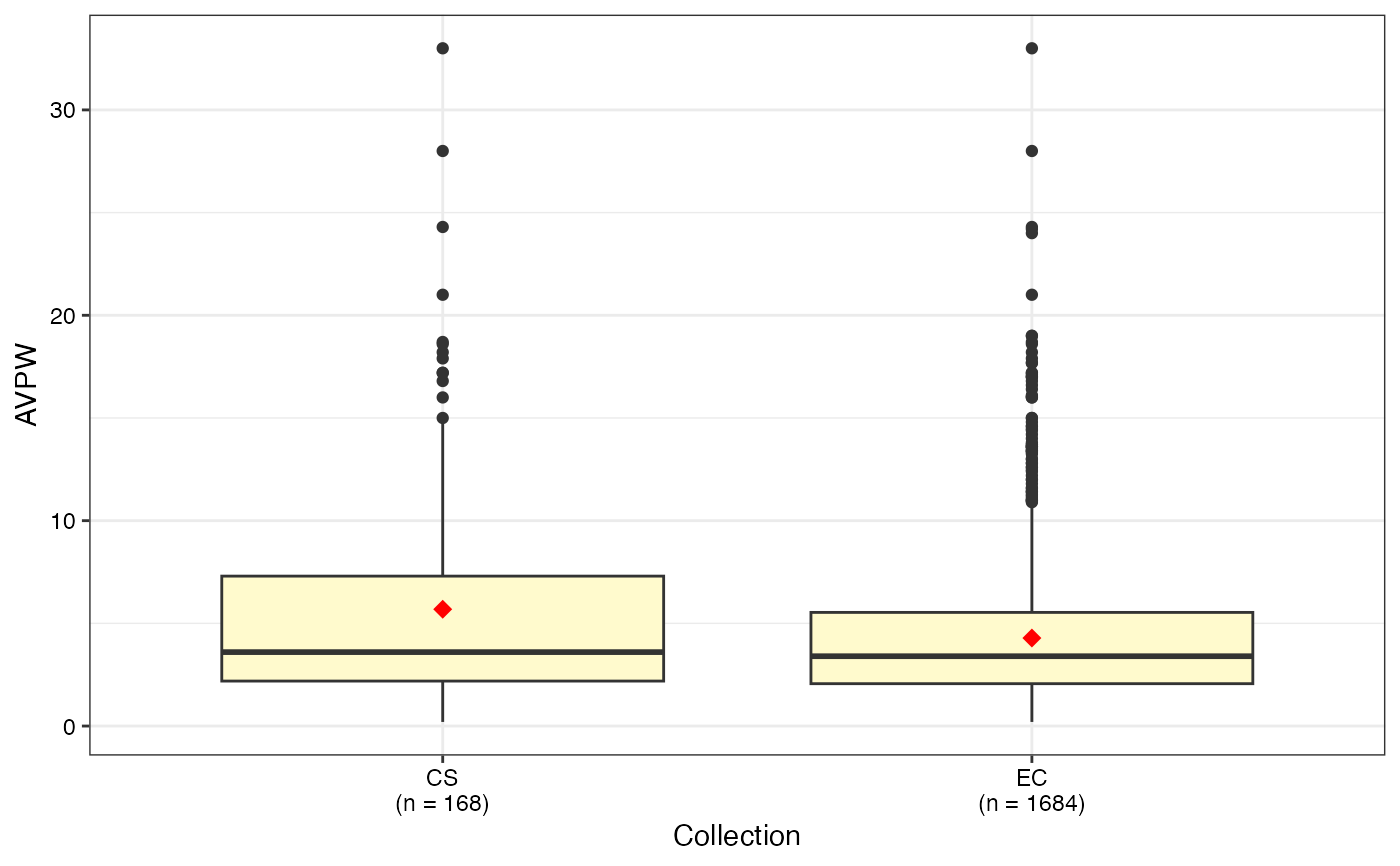 #>
#> $ARSR
#>
#> $ARSR
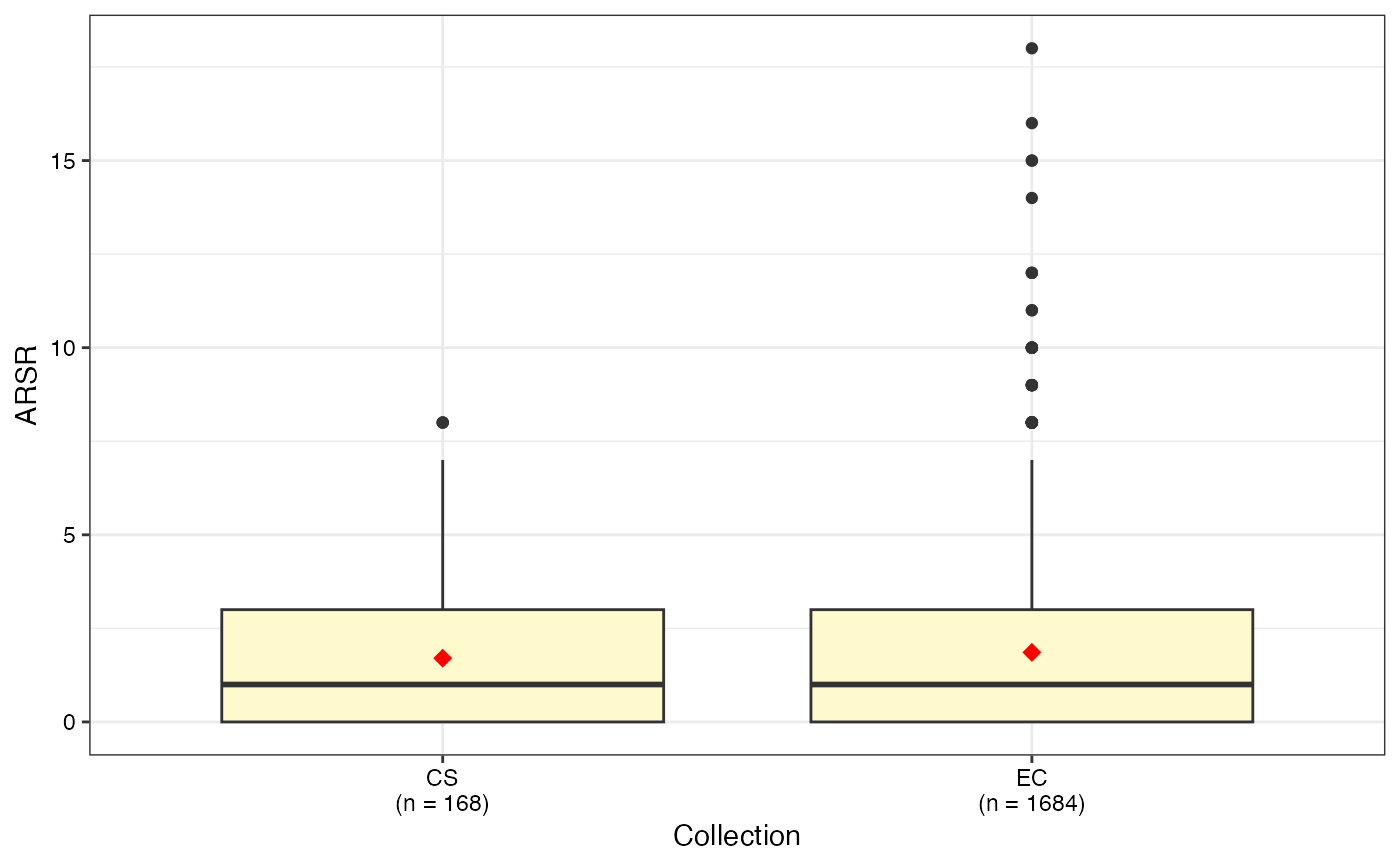 #>
#> $SRDM
#>
#> $SRDM
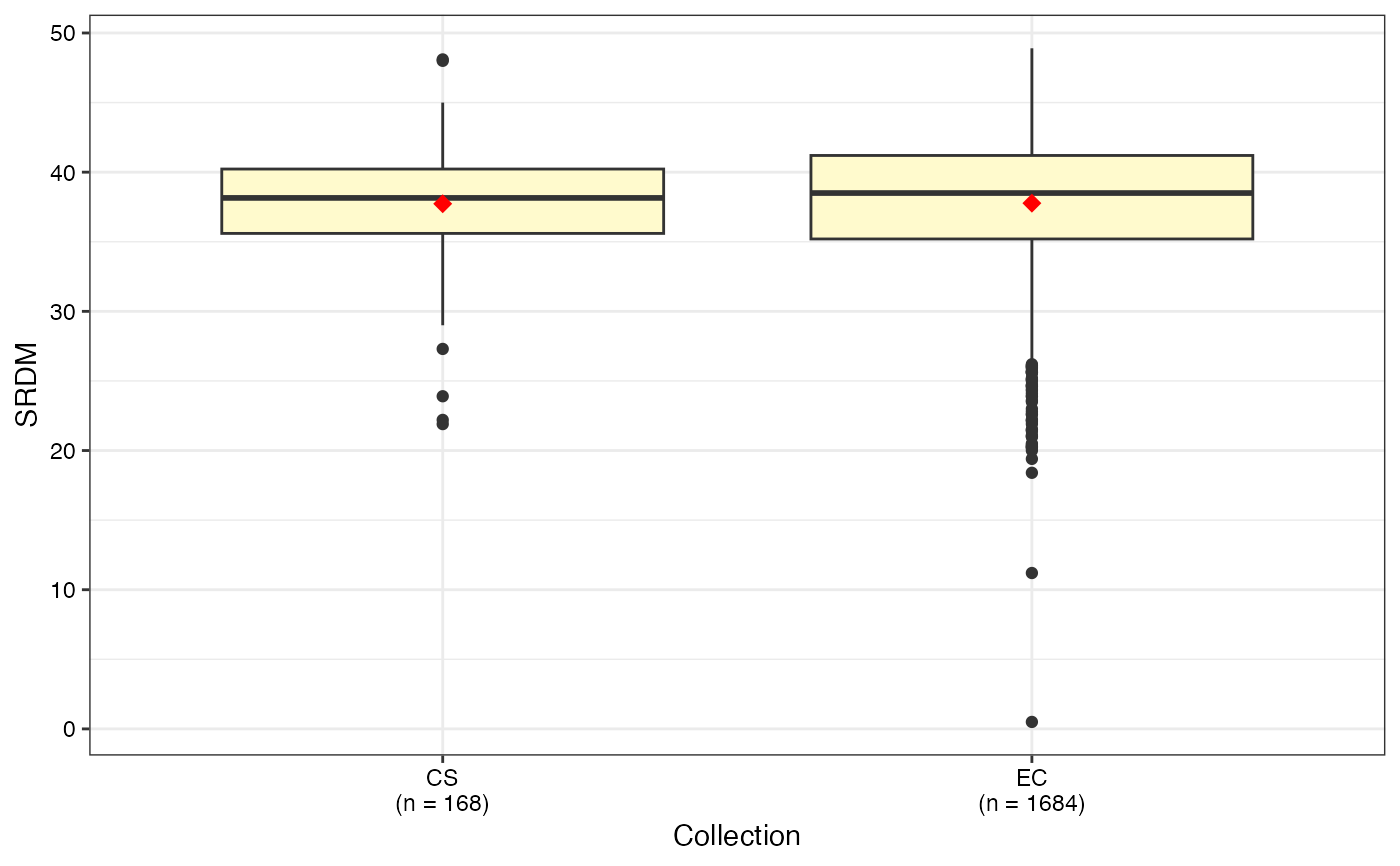 #>
# }
#>
# }