An example germplasm characterisation data of a subset of IITA Cassava
collection
(International Institute of Tropical Agriculture et al. 2019)
.
Includes data on 26 (out of 62) descriptors for 1684 (out of 2170)
accessions. It is used to demonstrate the various functions of
EvaluateCore package.
Format
A data frame with 58 columns:
- CUAL
Colour of unexpanded apical leaves
- LNGS
Length of stipules
- PTLC
Petiole colour
- DSTA
Distribution of anthocyanin
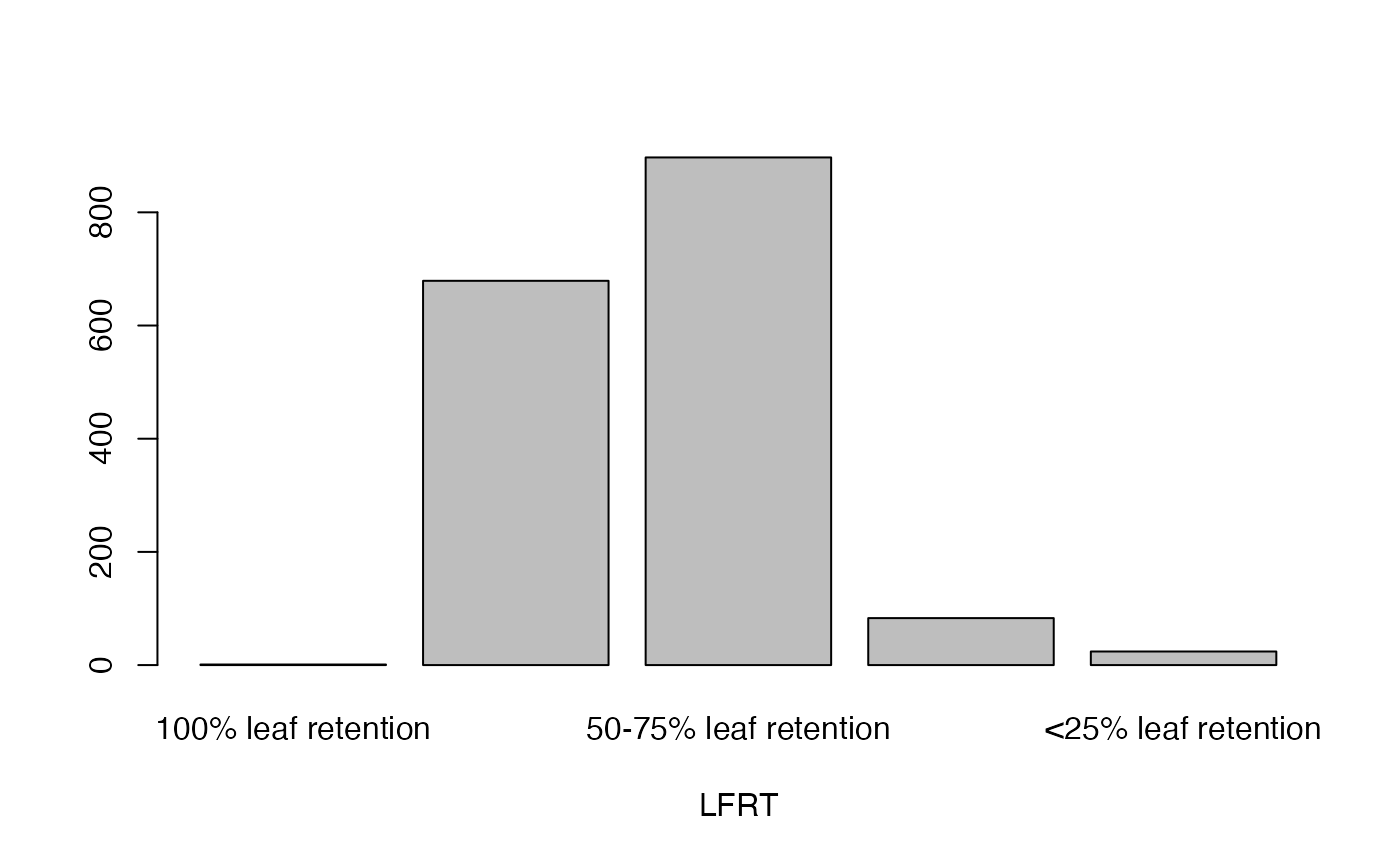
- LFRT
Leaf retention
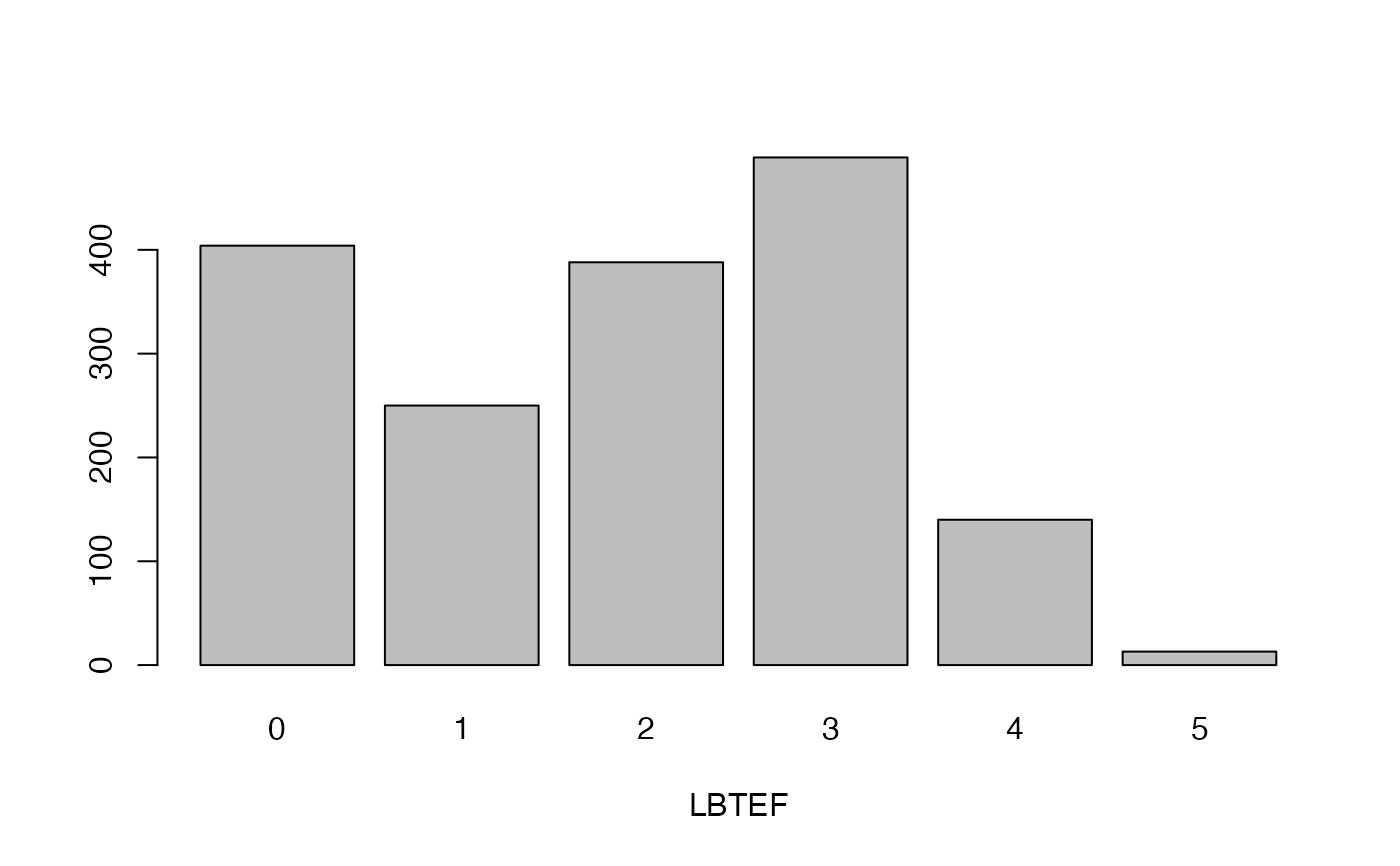
- LBTEF
Level of branching at the end of flowering
- CBTR
Colour of boiled tuberous root
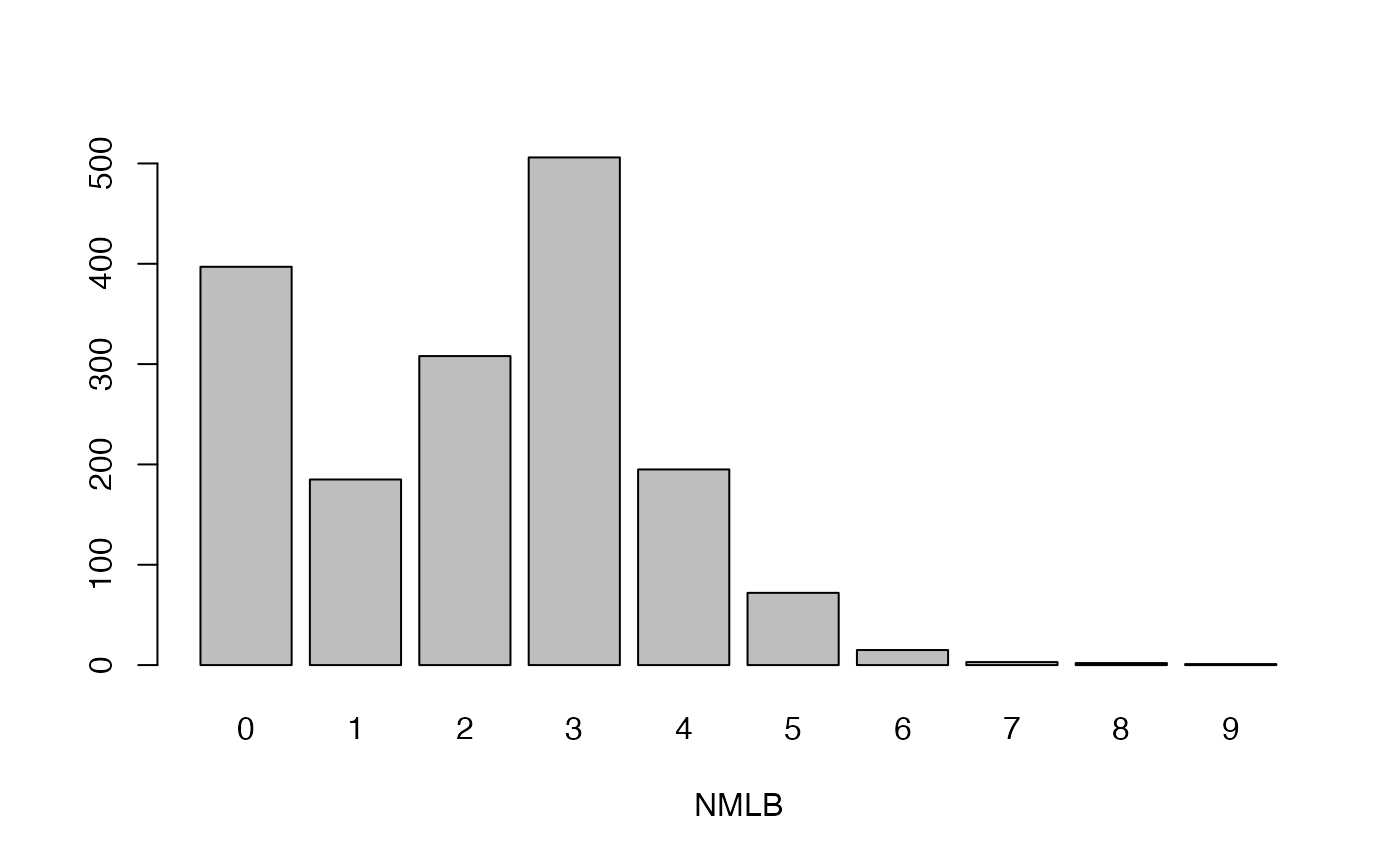
- NMLB
Number of levels of branching
- ANGB
Angle of branching
- CUAL9M
Colours of unexpanded apical leaves at 9 months
- LVC9M
Leaf vein colour at 9 months
- TNPR9M
Total number of plants remaining per accession at 9 months
- PL9M
Petiole length at 9 months
- STRP
Storage root peduncle
- STRC
Storage root constrictions
- PSTR
Position of root
- NMSR
Number of storage root per plant
- TTRN
Total root number per plant
- TFWSR
Total fresh weight of storage root per plant
- TTRW
Total root weight per plant
- TFWSS
Total fresh weight of storage shoot per plant
- TTSW
Total shoot weight per plant
- TTPW
Total plant weight
- AVPW
Average plant weight
- ARSR
Amount of rotted storage root per plant
- SRDM
Storage root dry matter
Details
Further details on how the example dataset was built from the original data is available online.
References
International Institute of Tropical Agriculture, Benjamin F, Marimagne T (2019). “Cassava morphological characterization. Version 2018.1.”
Examples
data(cassava_EC)
summary(cassava_EC)
#> CUAL LNGS PTLC DSTA
#> Length:1684 Length:1684 Length:1684 Length:1684
#> Class :character Class :character Class :character Class :character
#> Mode :character Mode :character Mode :character Mode :character
#>
#>
#>
#> LFRT LBTEF CBTR NMLB
#> Length:1684 Length:1684 Length:1684 Length:1684
#> Class :character Class :character Class :character Class :character
#> Mode :character Mode :character Mode :character Mode :character
#>
#>
#>
#> ANGB CUAL9M LVC9M TNPR9M
#> Length:1684 Length:1684 Length:1684 Length:1684
#> Class :character Class :character Class :character Class :character
#> Mode :character Mode :character Mode :character Mode :character
#>
#>
#>
#> PL9M STRP STRC PSTR
#> Length:1684 Length:1684 Length:1684 Length:1684
#> Class :character Class :character Class :character Class :character
#> Mode :character Mode :character Mode :character Mode :character
#>
#>
#>
#> NMSR TTRN TFWSR TTRW
#> Min. : 1.00 Min. : 0.250 Min. : 0.000 Min. : 0.000
#> 1st Qu.: 6.00 1st Qu.: 2.500 1st Qu.: 2.200 1st Qu.: 0.900
#> Median :10.00 Median : 3.600 Median : 4.200 Median : 1.445
#> Mean :11.72 Mean : 3.854 Mean : 5.429 Mean : 1.898
#> 3rd Qu.:16.00 3rd Qu.: 5.000 3rd Qu.: 7.000 3rd Qu.: 2.400
#> Max. :55.00 Max. :13.750 Max. :40.000 Max. :20.200
#> TFWSS TTSW TTPW AVPW
#> Min. : 0.200 Min. : 0.040 Min. : 0.40 Min. : 0.200
#> 1st Qu.: 2.600 1st Qu.: 1.000 1st Qu.: 5.20 1st Qu.: 2.062
#> Median : 5.400 Median : 1.933 Median :10.00 Median : 3.400
#> Mean : 6.943 Mean : 2.388 Mean :12.37 Mean : 4.285
#> 3rd Qu.:10.000 3rd Qu.: 3.200 3rd Qu.:16.45 3rd Qu.: 5.533
#> Max. :42.000 Max. :22.000 Max. :80.00 Max. :33.000
#> ARSR SRDM
#> Min. : 0.000 Min. : 0.50
#> 1st Qu.: 0.000 1st Qu.:35.20
#> Median : 1.000 Median :38.50
#> Mean : 1.858 Mean :37.77
#> 3rd Qu.: 3.000 3rd Qu.:41.20
#> Max. :18.000 Max. :48.90
quant <- c("NMSR", "TTRN", "TFWSR", "TTRW", "TFWSS", "TTSW", "TTPW", "AVPW",
"ARSR", "SRDM")
qual <- c("CUAL", "LNGS", "PTLC", "DSTA", "LFRT", "LBTEF", "CBTR", "NMLB",
"ANGB", "CUAL9M", "LVC9M", "TNPR9M", "PL9M", "STRP", "STRC",
"PSTR")
lapply(seq_along(cassava_EC[, qual]),
function(i) barplot(table(cassava_EC[, qual][, i]),
xlab = names(cassava_EC[, qual])[i]))
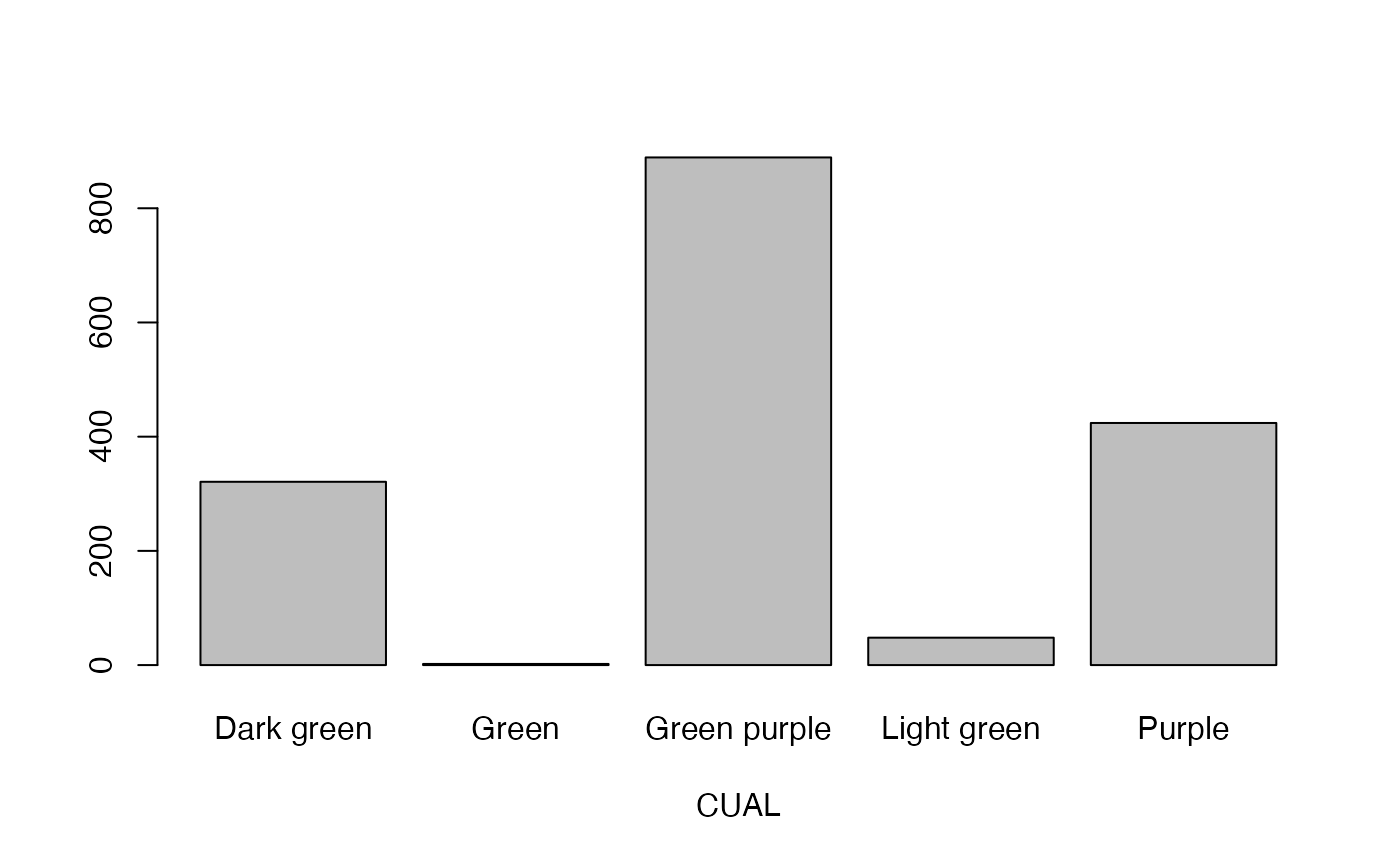
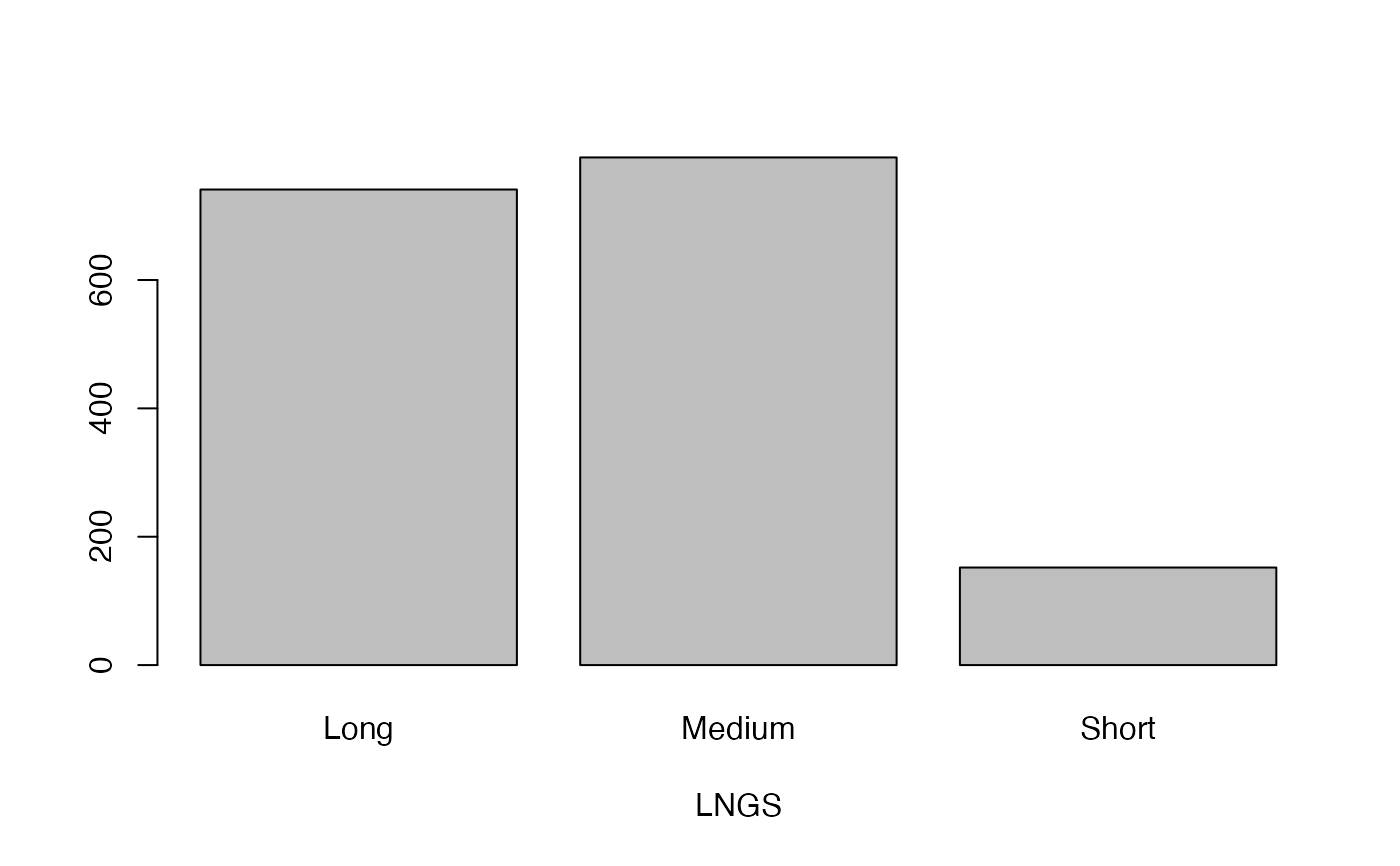
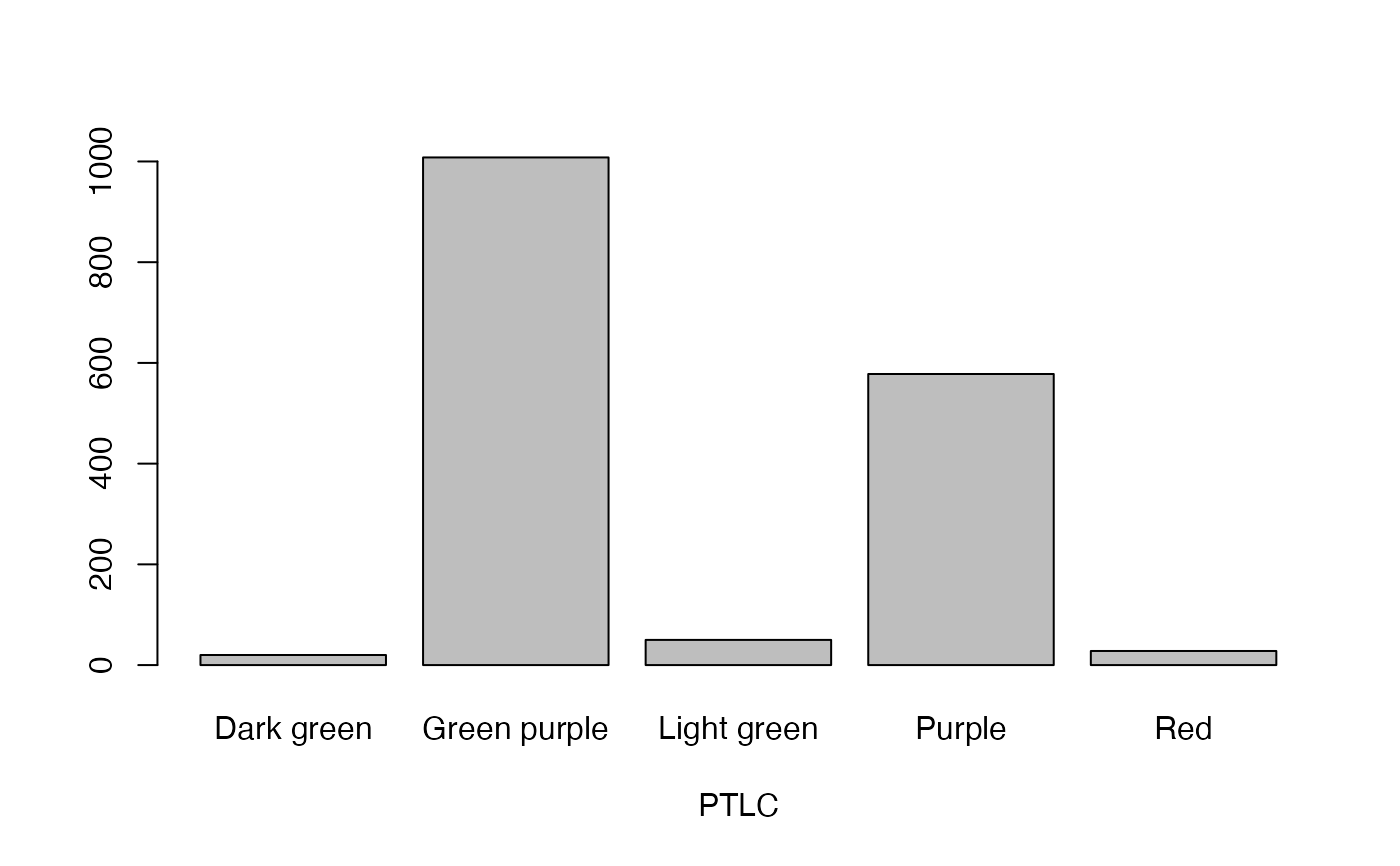

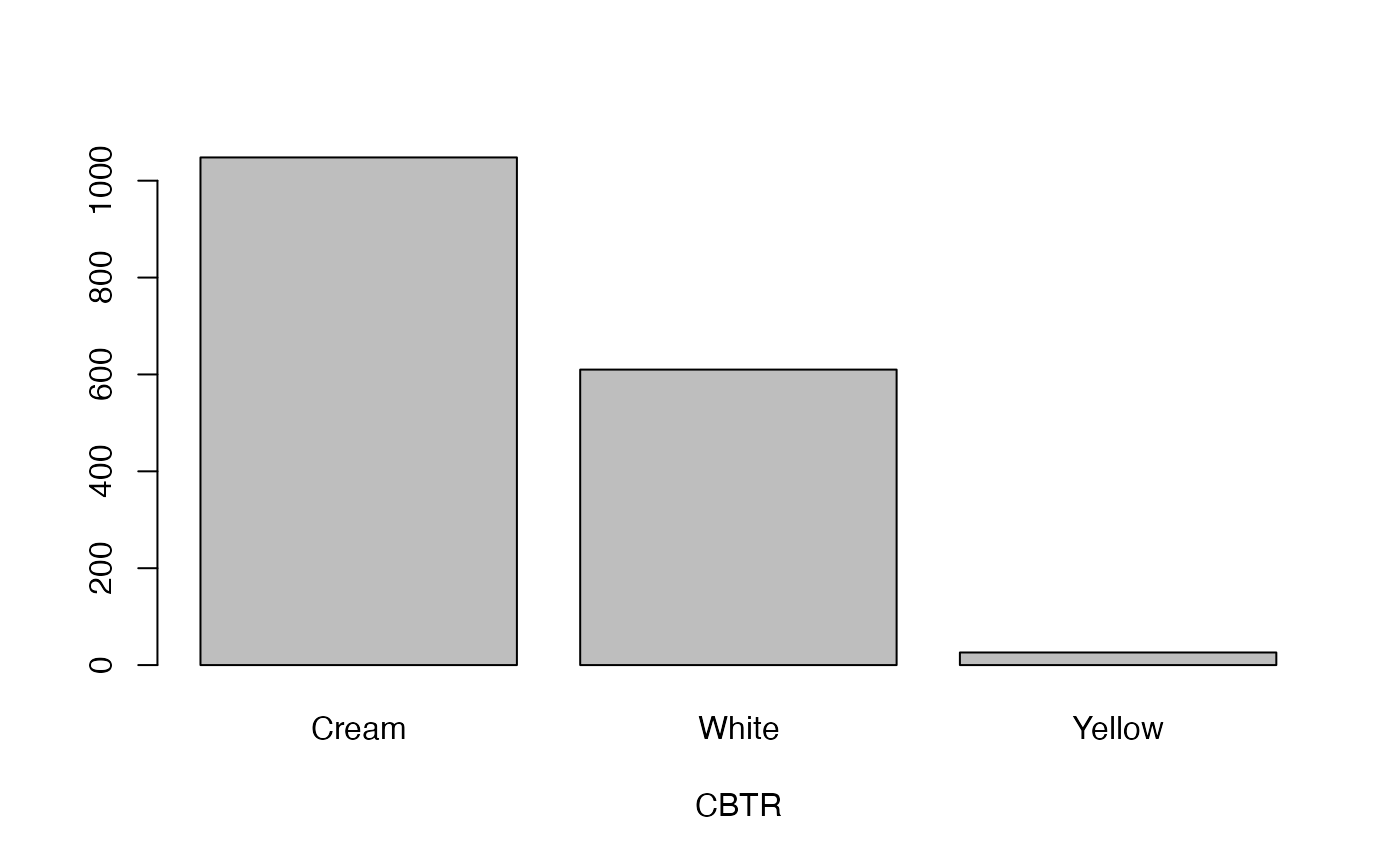
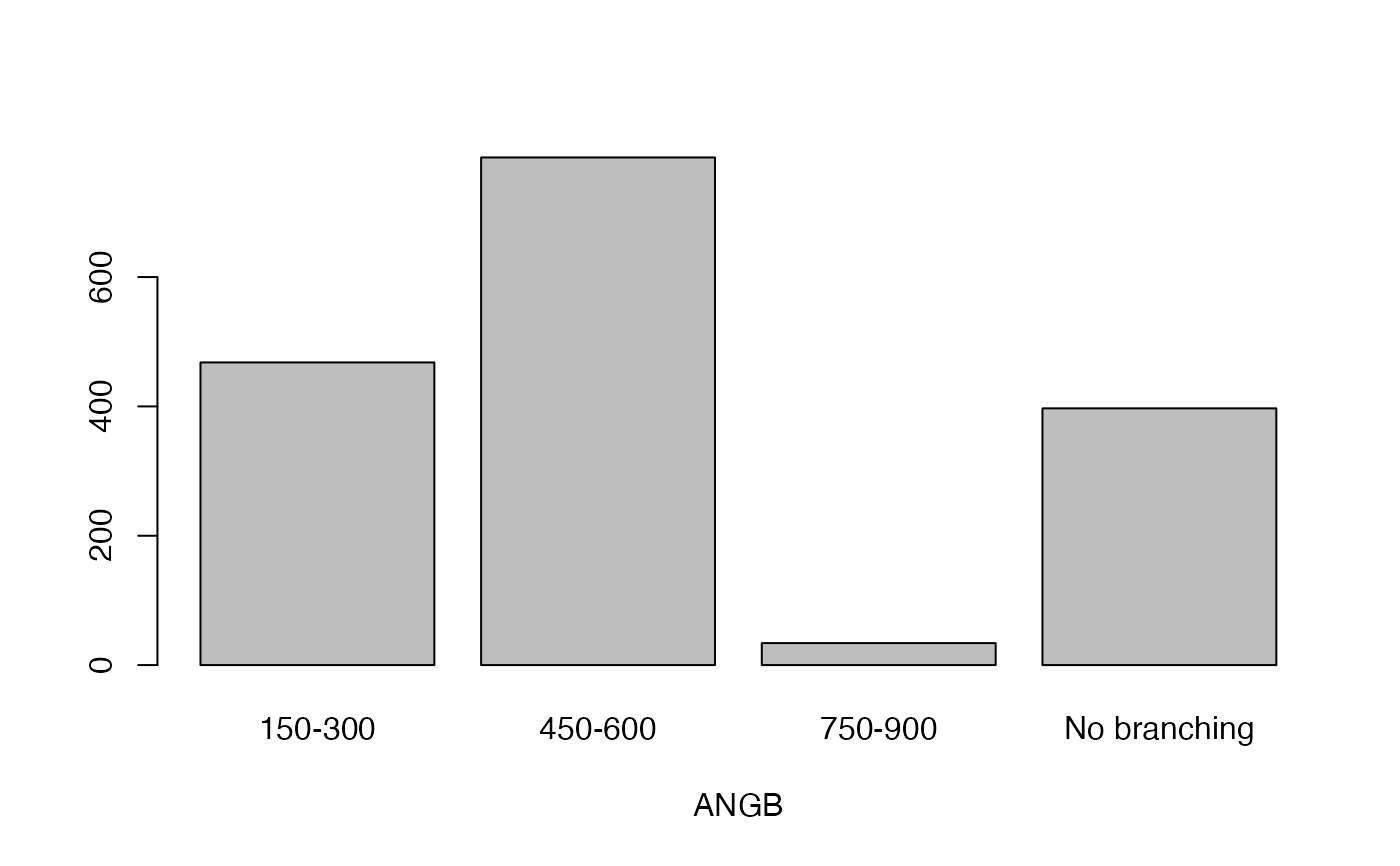
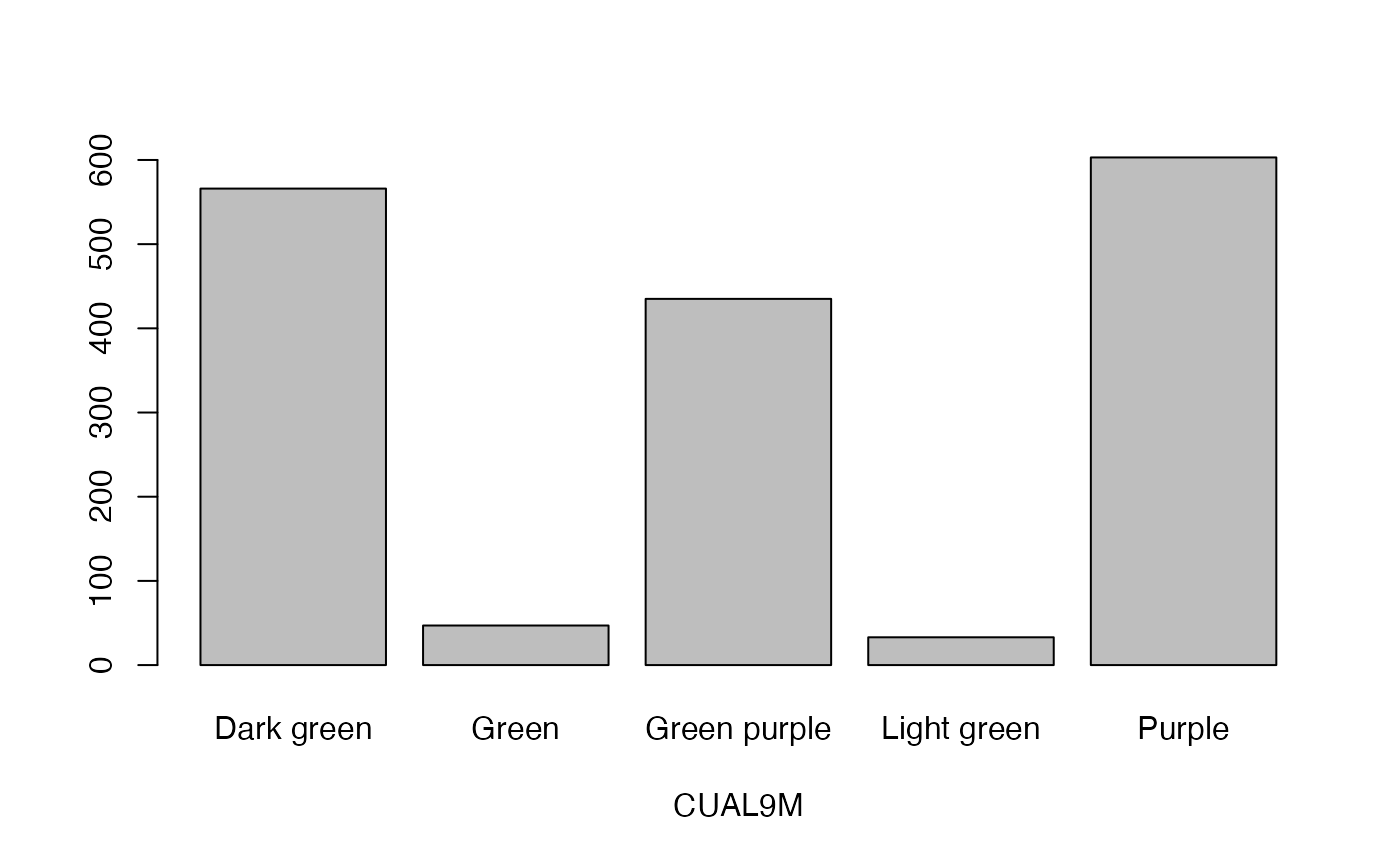
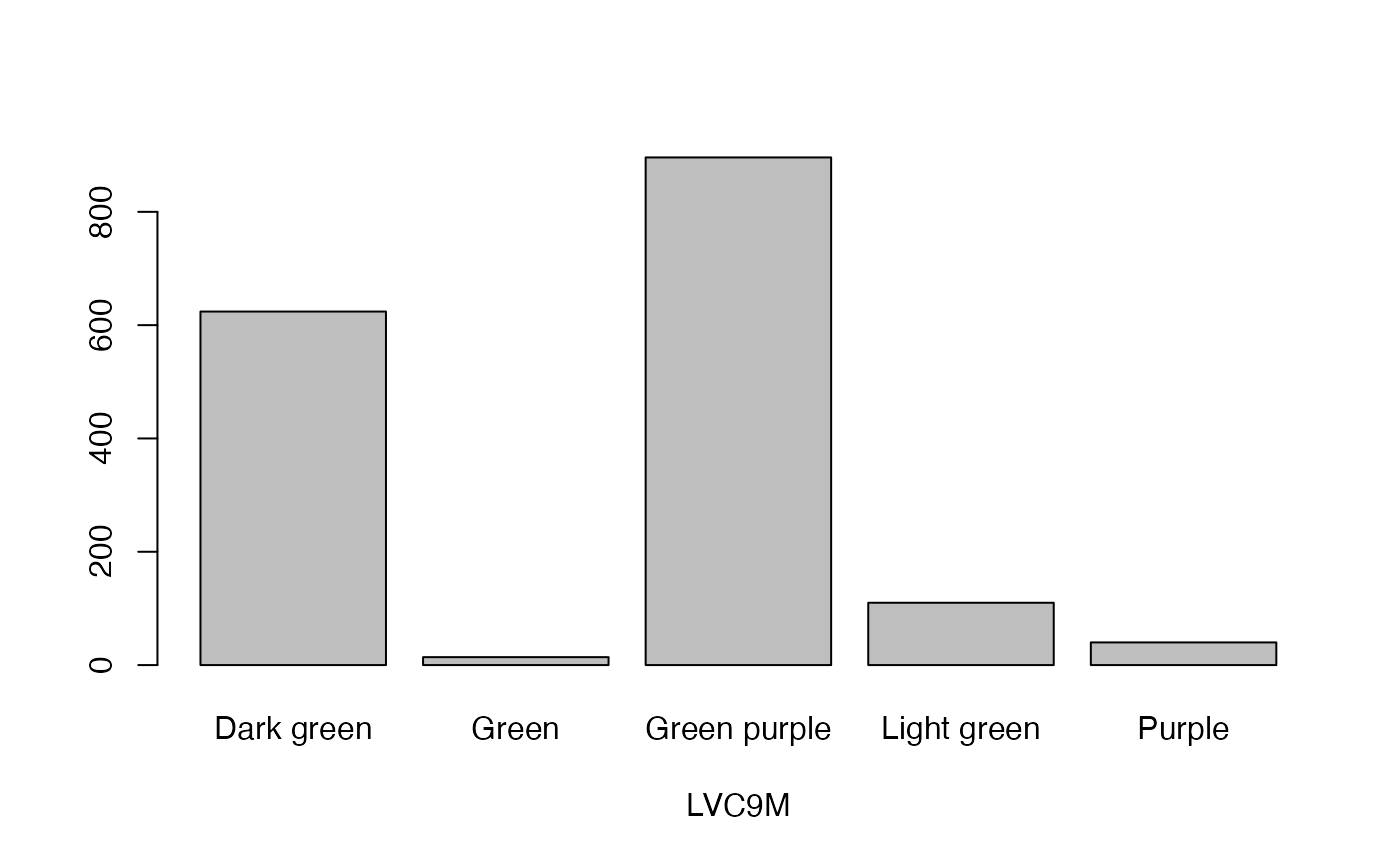
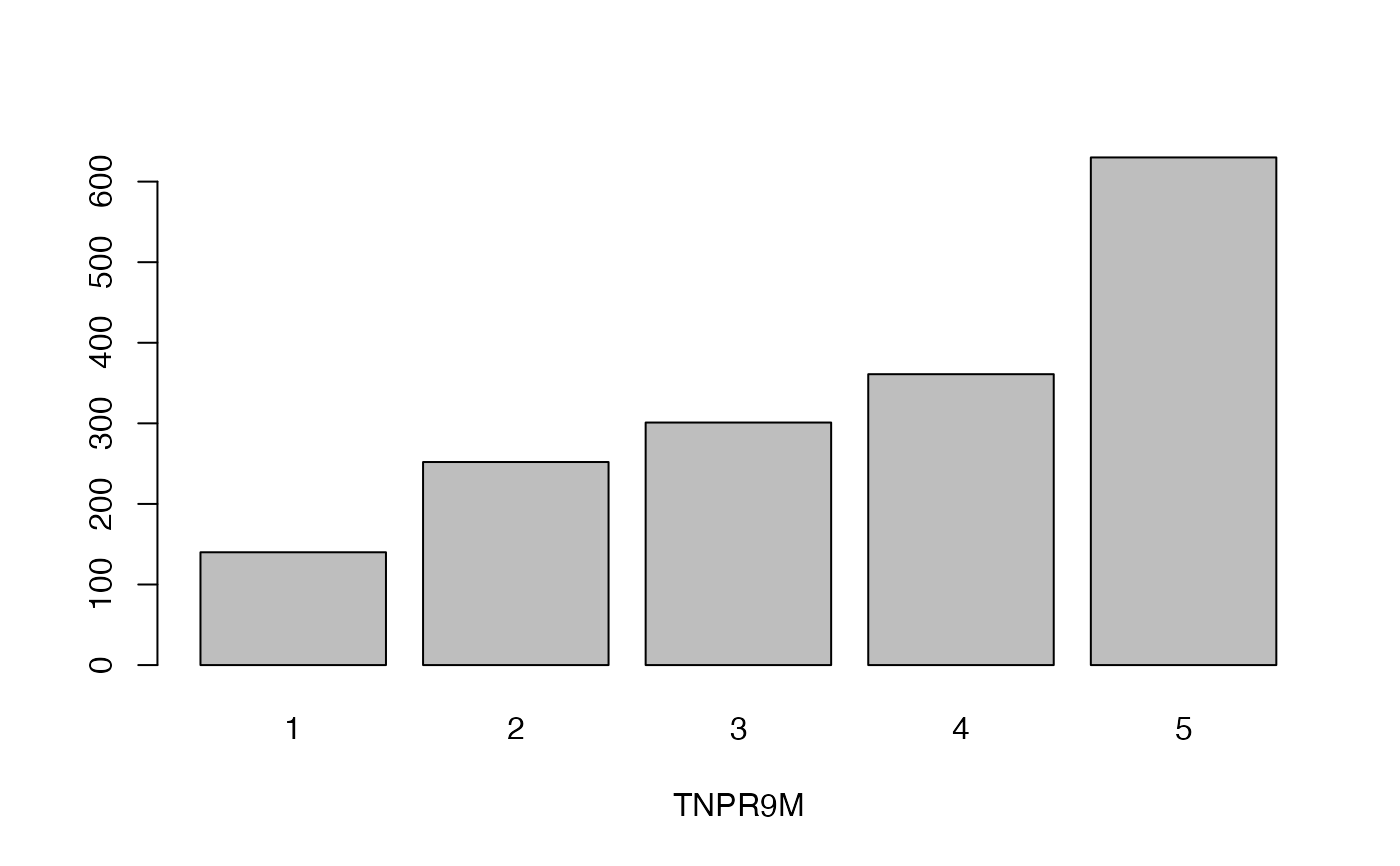
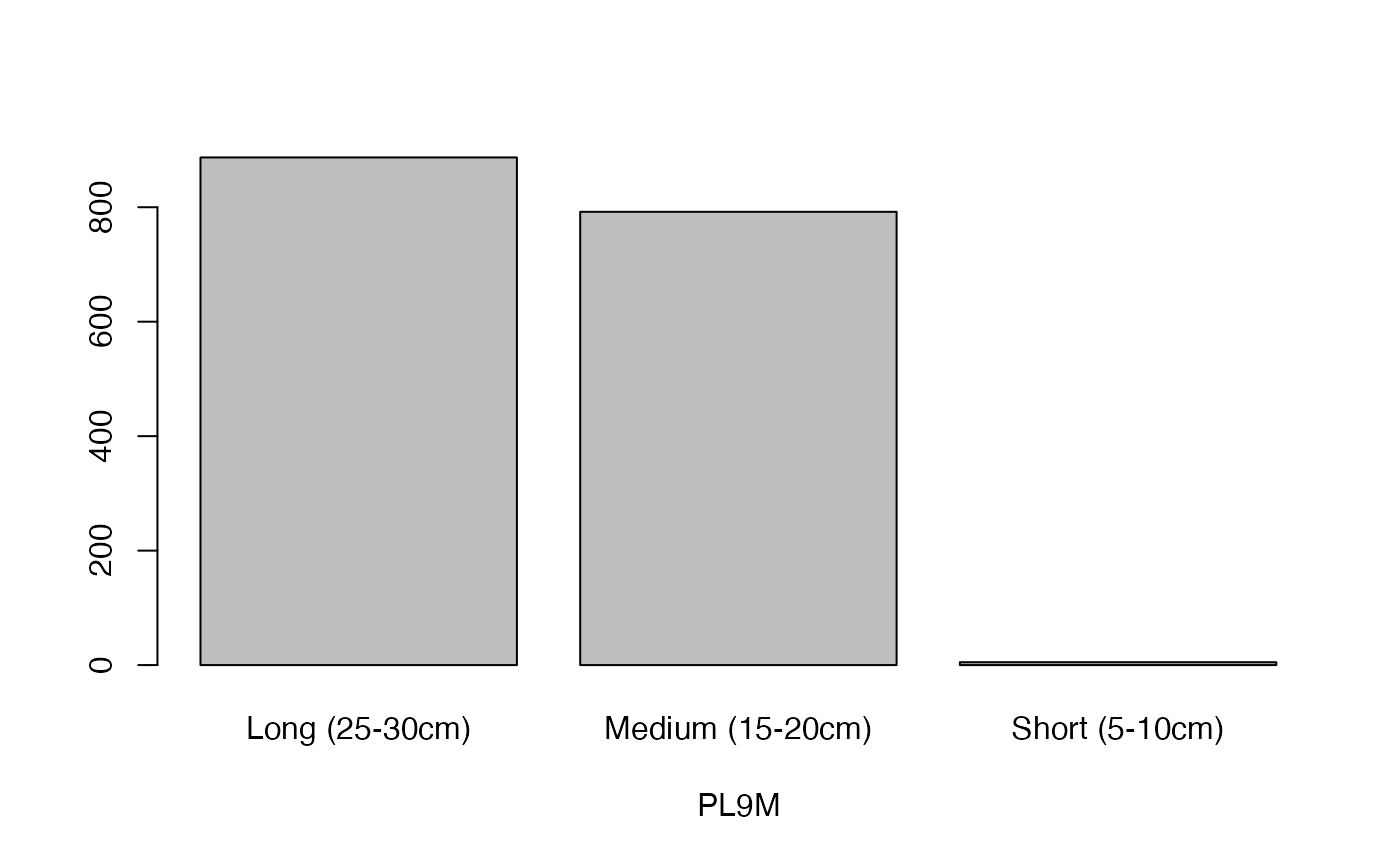
 #> [[1]]
#> [,1]
#> [1,] 0.7
#> [2,] 1.9
#> [3,] 3.1
#> [4,] 4.3
#> [5,] 5.5
#>
#> [[2]]
#> [,1]
#> [1,] 0.7
#> [2,] 1.9
#> [3,] 3.1
#>
#> [[3]]
#> [,1]
#> [1,] 0.7
#> [2,] 1.9
#> [3,] 3.1
#> [4,] 4.3
#> [5,] 5.5
#>
#> [[4]]
#> [,1]
#> [1,] 0.7
#> [2,] 1.9
#> [3,] 3.1
#> [4,] 4.3
#> [5,] 5.5
#>
#> [[5]]
#> [,1]
#> [1,] 0.7
#> [2,] 1.9
#> [3,] 3.1
#> [4,] 4.3
#> [5,] 5.5
#>
#> [[6]]
#> [,1]
#> [1,] 0.7
#> [2,] 1.9
#> [3,] 3.1
#> [4,] 4.3
#> [5,] 5.5
#> [6,] 6.7
#>
#> [[7]]
#> [,1]
#> [1,] 0.7
#> [2,] 1.9
#> [3,] 3.1
#>
#> [[8]]
#> [,1]
#> [1,] 0.7
#> [2,] 1.9
#> [3,] 3.1
#> [4,] 4.3
#> [5,] 5.5
#> [6,] 6.7
#> [7,] 7.9
#> [8,] 9.1
#> [9,] 10.3
#> [10,] 11.5
#>
#> [[9]]
#> [,1]
#> [1,] 0.7
#> [2,] 1.9
#> [3,] 3.1
#> [4,] 4.3
#>
#> [[10]]
#> [,1]
#> [1,] 0.7
#> [2,] 1.9
#> [3,] 3.1
#> [4,] 4.3
#> [5,] 5.5
#>
#> [[11]]
#> [,1]
#> [1,] 0.7
#> [2,] 1.9
#> [3,] 3.1
#> [4,] 4.3
#> [5,] 5.5
#>
#> [[12]]
#> [,1]
#> [1,] 0.7
#> [2,] 1.9
#> [3,] 3.1
#> [4,] 4.3
#> [5,] 5.5
#>
#> [[13]]
#> [,1]
#> [1,] 0.7
#> [2,] 1.9
#> [3,] 3.1
#>
#> [[14]]
#> [,1]
#> [1,] 0.7
#> [2,] 1.9
#> [3,] 3.1
#> [4,] 4.3
#>
#> [[15]]
#> [,1]
#> [1,] 0.7
#> [2,] 1.9
#>
#> [[16]]
#> [,1]
#> [1,] 0.7
#> [2,] 1.9
#> [3,] 3.1
#>
lapply(seq_along(cassava_EC[, quant]),
function(i) hist(table(cassava_EC[, quant][, i]),
xlab = names(cassava_EC[, quant])[i],
main = ""))
#> [[1]]
#> [,1]
#> [1,] 0.7
#> [2,] 1.9
#> [3,] 3.1
#> [4,] 4.3
#> [5,] 5.5
#>
#> [[2]]
#> [,1]
#> [1,] 0.7
#> [2,] 1.9
#> [3,] 3.1
#>
#> [[3]]
#> [,1]
#> [1,] 0.7
#> [2,] 1.9
#> [3,] 3.1
#> [4,] 4.3
#> [5,] 5.5
#>
#> [[4]]
#> [,1]
#> [1,] 0.7
#> [2,] 1.9
#> [3,] 3.1
#> [4,] 4.3
#> [5,] 5.5
#>
#> [[5]]
#> [,1]
#> [1,] 0.7
#> [2,] 1.9
#> [3,] 3.1
#> [4,] 4.3
#> [5,] 5.5
#>
#> [[6]]
#> [,1]
#> [1,] 0.7
#> [2,] 1.9
#> [3,] 3.1
#> [4,] 4.3
#> [5,] 5.5
#> [6,] 6.7
#>
#> [[7]]
#> [,1]
#> [1,] 0.7
#> [2,] 1.9
#> [3,] 3.1
#>
#> [[8]]
#> [,1]
#> [1,] 0.7
#> [2,] 1.9
#> [3,] 3.1
#> [4,] 4.3
#> [5,] 5.5
#> [6,] 6.7
#> [7,] 7.9
#> [8,] 9.1
#> [9,] 10.3
#> [10,] 11.5
#>
#> [[9]]
#> [,1]
#> [1,] 0.7
#> [2,] 1.9
#> [3,] 3.1
#> [4,] 4.3
#>
#> [[10]]
#> [,1]
#> [1,] 0.7
#> [2,] 1.9
#> [3,] 3.1
#> [4,] 4.3
#> [5,] 5.5
#>
#> [[11]]
#> [,1]
#> [1,] 0.7
#> [2,] 1.9
#> [3,] 3.1
#> [4,] 4.3
#> [5,] 5.5
#>
#> [[12]]
#> [,1]
#> [1,] 0.7
#> [2,] 1.9
#> [3,] 3.1
#> [4,] 4.3
#> [5,] 5.5
#>
#> [[13]]
#> [,1]
#> [1,] 0.7
#> [2,] 1.9
#> [3,] 3.1
#>
#> [[14]]
#> [,1]
#> [1,] 0.7
#> [2,] 1.9
#> [3,] 3.1
#> [4,] 4.3
#>
#> [[15]]
#> [,1]
#> [1,] 0.7
#> [2,] 1.9
#>
#> [[16]]
#> [,1]
#> [1,] 0.7
#> [2,] 1.9
#> [3,] 3.1
#>
lapply(seq_along(cassava_EC[, quant]),
function(i) hist(table(cassava_EC[, quant][, i]),
xlab = names(cassava_EC[, quant])[i],
main = ""))
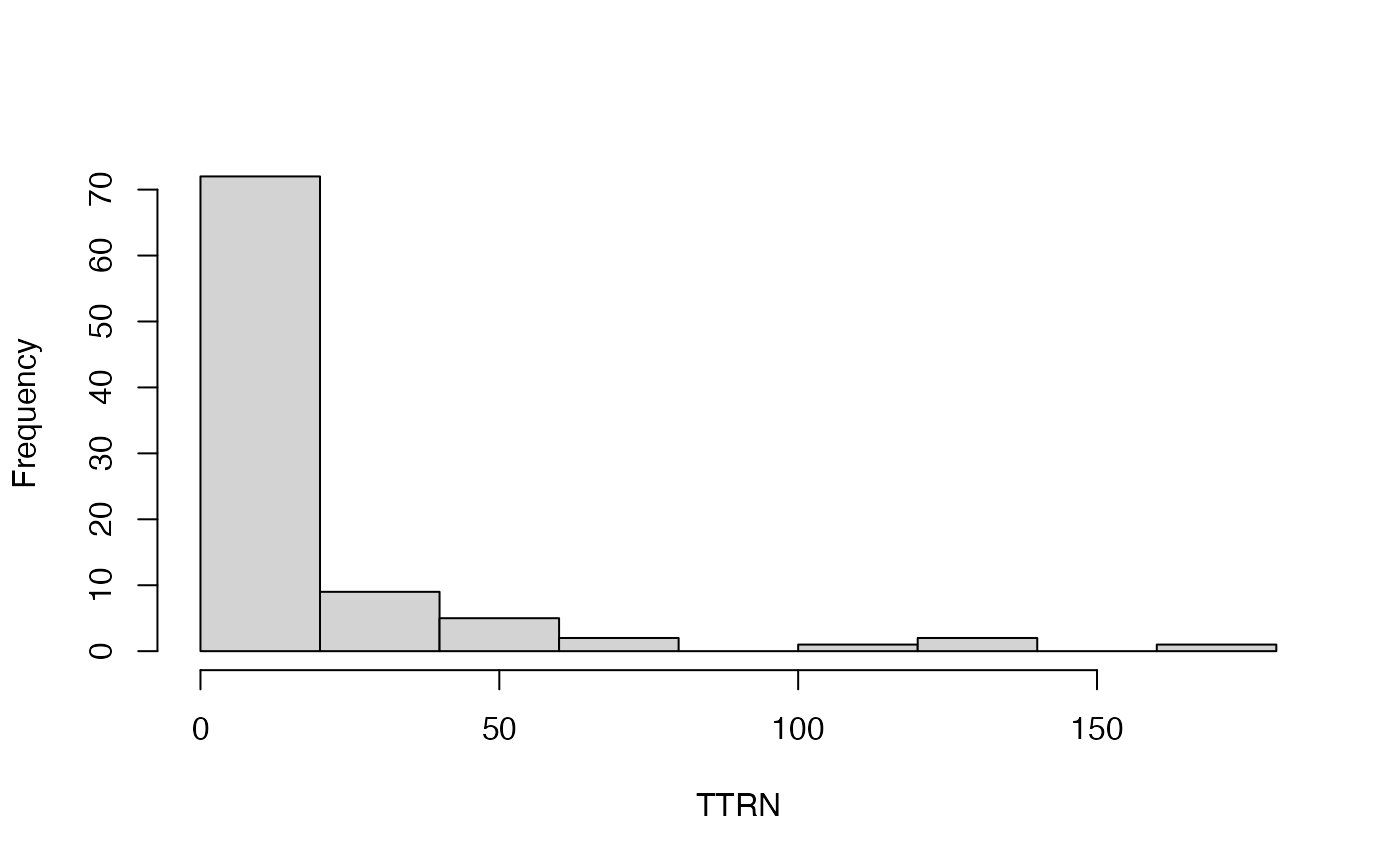
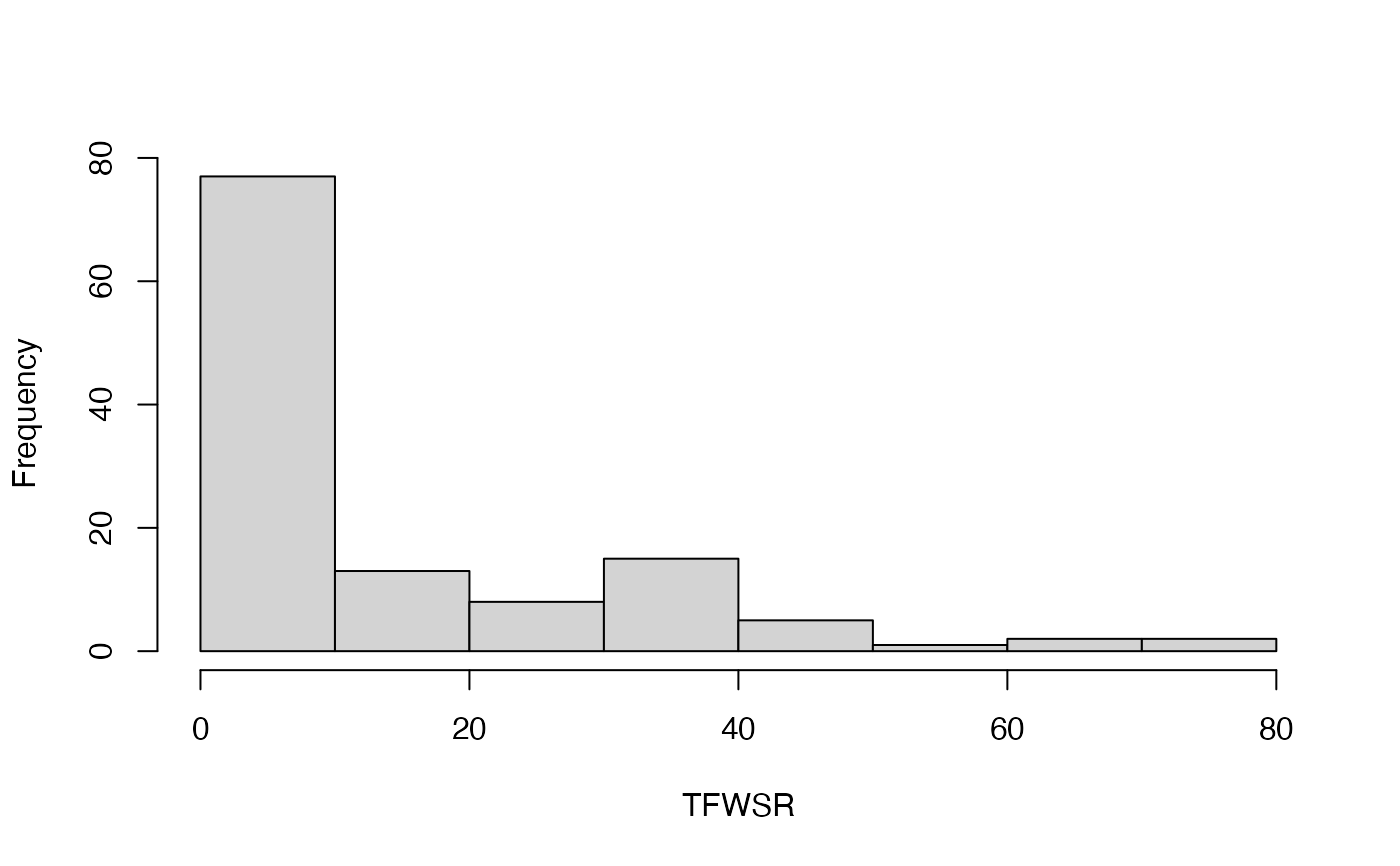
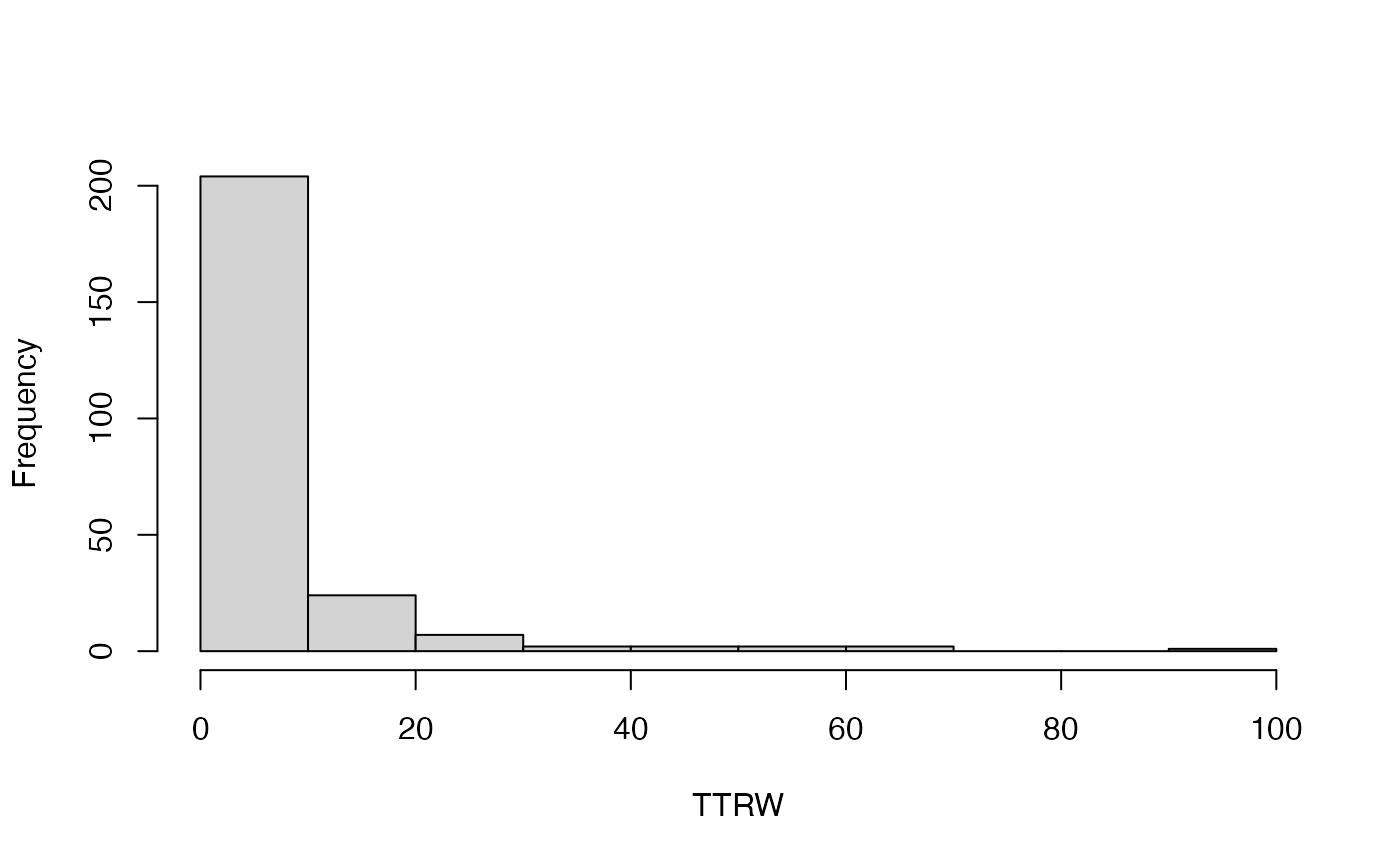
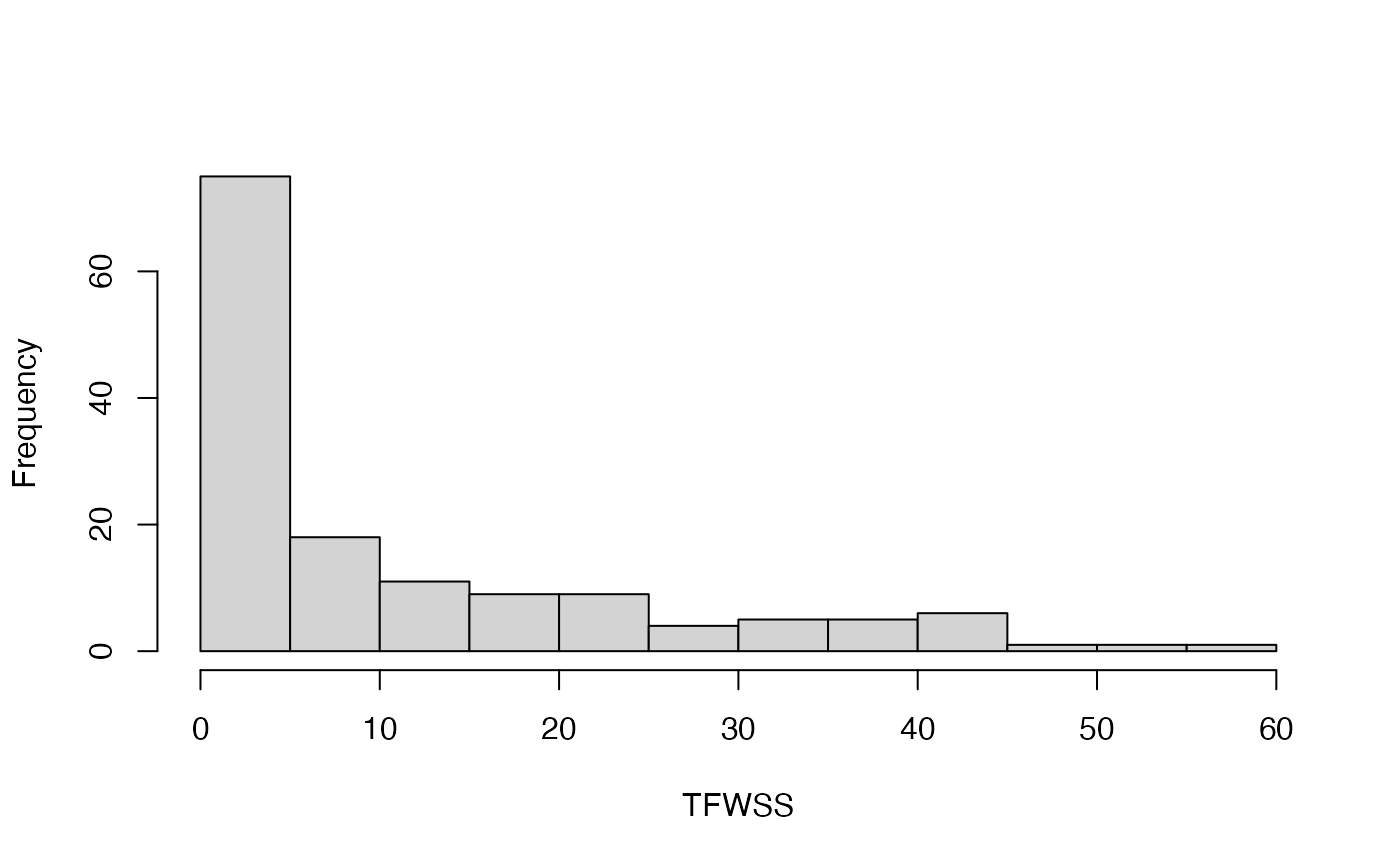
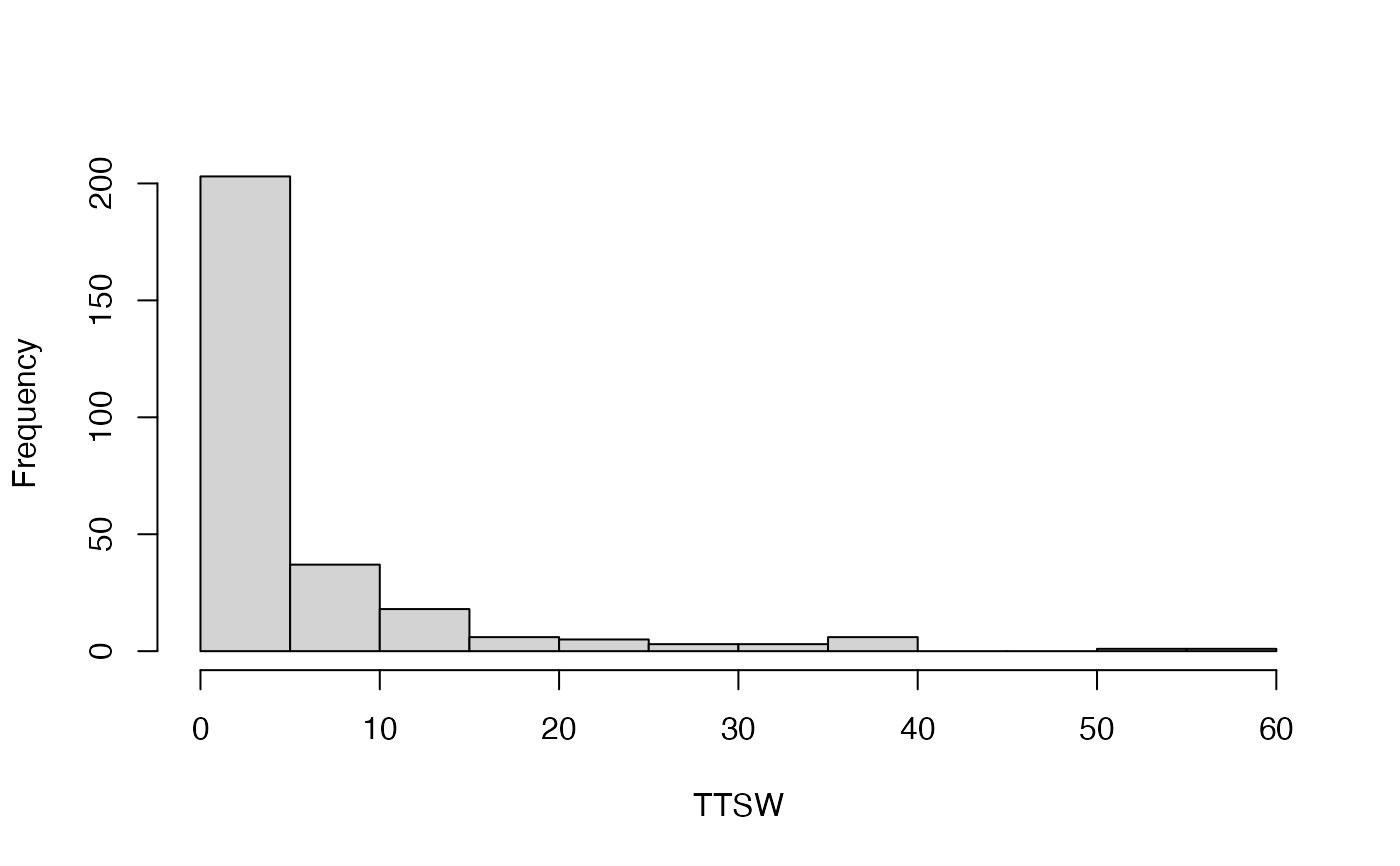
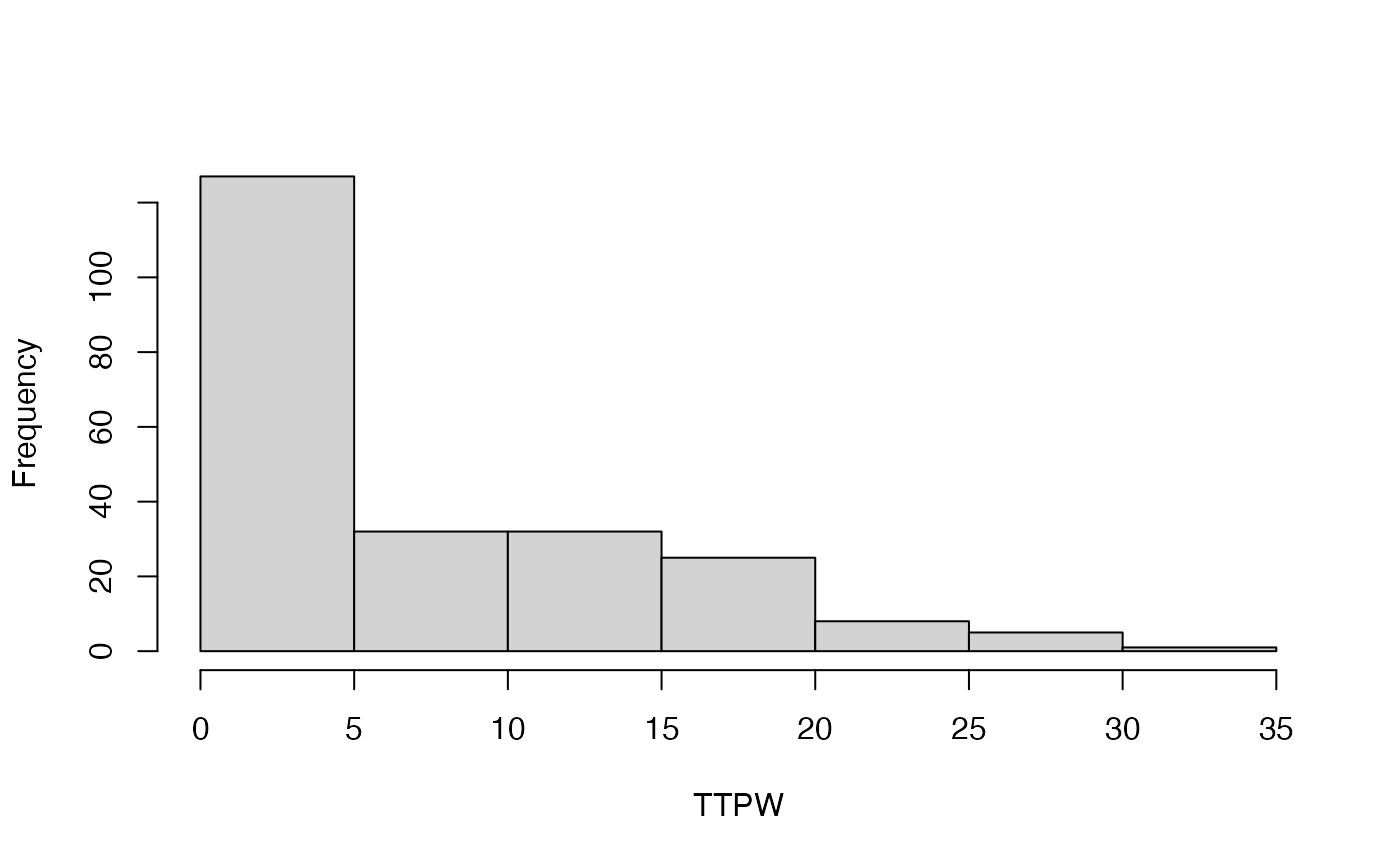
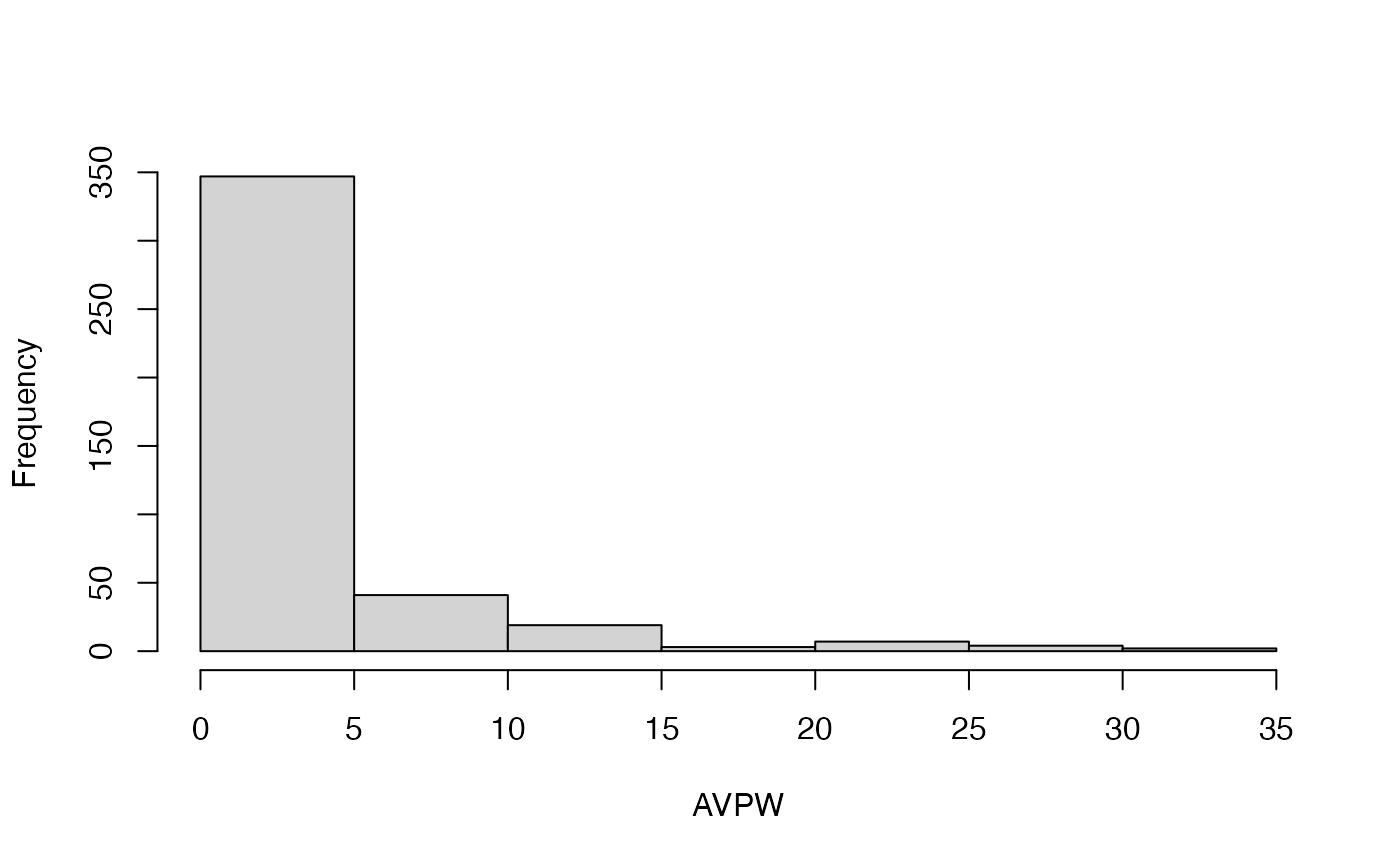
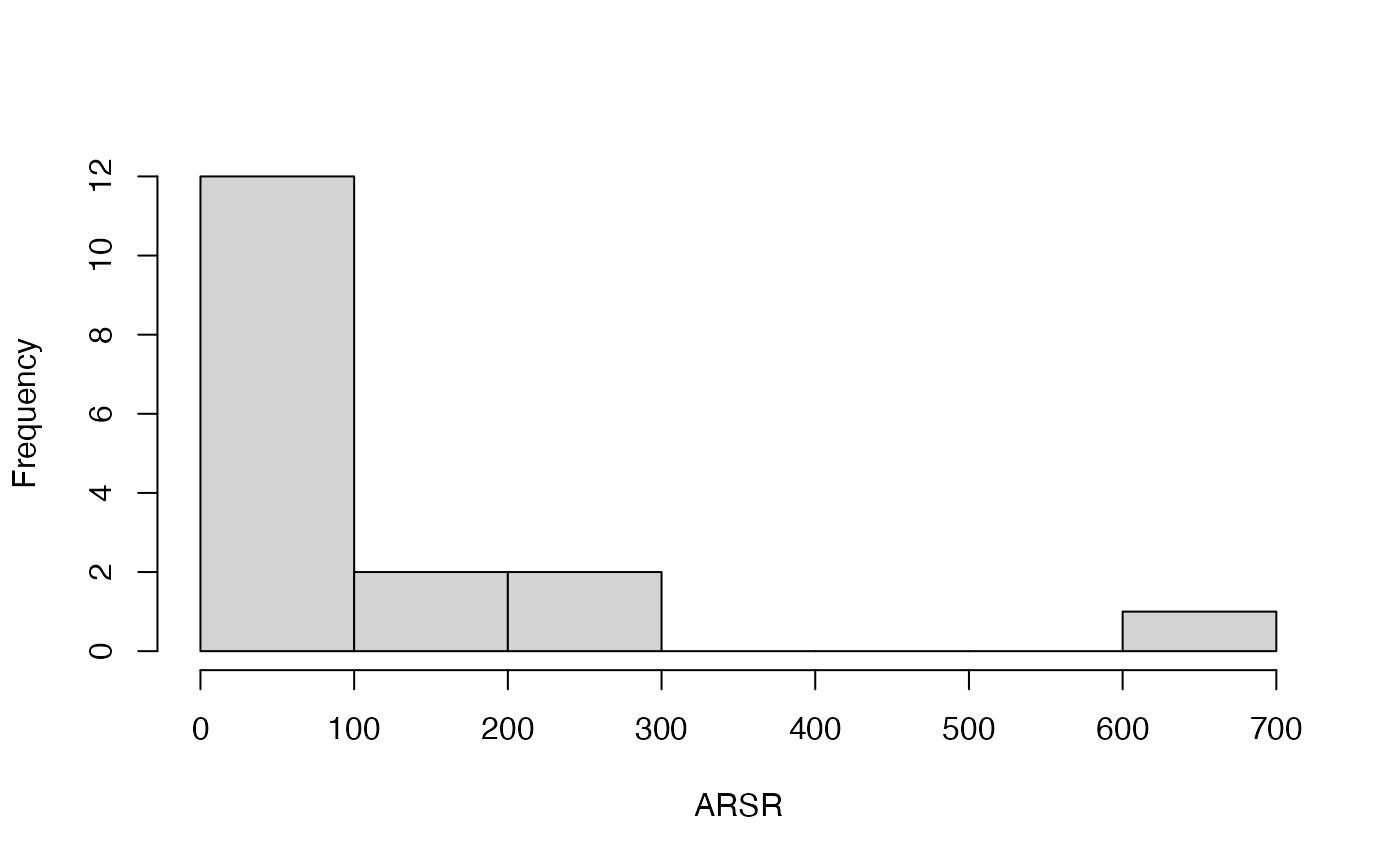
 #> [[1]]
#> $breaks
#> [1] 0 20 40 60 80 100 120
#>
#> $counts
#> [1] 18 5 5 6 7 1
#>
#> $density
#> [1] 0.021428571 0.005952381 0.005952381 0.007142857 0.008333333 0.001190476
#>
#> $mids
#> [1] 10 30 50 70 90 110
#>
#> $xname
#> [1] "table(cassava_EC[, quant][, i])"
#>
#> $equidist
#> [1] TRUE
#>
#> attr(,"class")
#> [1] "histogram"
#>
#> [[2]]
#> $breaks
#> [1] 0 20 40 60 80 100 120 140 160 180
#>
#> $counts
#> [1] 72 9 5 2 0 1 2 0 1
#>
#> $density
#> [1] 0.0391304348 0.0048913043 0.0027173913 0.0010869565 0.0000000000
#> [6] 0.0005434783 0.0010869565 0.0000000000 0.0005434783
#>
#> $mids
#> [1] 10 30 50 70 90 110 130 150 170
#>
#> $xname
#> [1] "table(cassava_EC[, quant][, i])"
#>
#> $equidist
#> [1] TRUE
#>
#> attr(,"class")
#> [1] "histogram"
#>
#> [[3]]
#> $breaks
#> [1] 0 10 20 30 40 50 60 70 80
#>
#> $counts
#> [1] 77 13 8 15 5 1 2 2
#>
#> $density
#> [1] 0.0626016260 0.0105691057 0.0065040650 0.0121951220 0.0040650407
#> [6] 0.0008130081 0.0016260163 0.0016260163
#>
#> $mids
#> [1] 5 15 25 35 45 55 65 75
#>
#> $xname
#> [1] "table(cassava_EC[, quant][, i])"
#>
#> $equidist
#> [1] TRUE
#>
#> attr(,"class")
#> [1] "histogram"
#>
#> [[4]]
#> $breaks
#> [1] 0 10 20 30 40 50 60 70 80 90 100
#>
#> $counts
#> [1] 204 24 7 2 2 2 2 0 0 1
#>
#> $density
#> [1] 0.0836065574 0.0098360656 0.0028688525 0.0008196721 0.0008196721
#> [6] 0.0008196721 0.0008196721 0.0000000000 0.0000000000 0.0004098361
#>
#> $mids
#> [1] 5 15 25 35 45 55 65 75 85 95
#>
#> $xname
#> [1] "table(cassava_EC[, quant][, i])"
#>
#> $equidist
#> [1] TRUE
#>
#> attr(,"class")
#> [1] "histogram"
#>
#> [[5]]
#> $breaks
#> [1] 0 5 10 15 20 25 30 35 40 45 50 55 60
#>
#> $counts
#> [1] 75 18 11 9 9 4 5 5 6 1 1 1
#>
#> $density
#> [1] 0.103448276 0.024827586 0.015172414 0.012413793 0.012413793 0.005517241
#> [7] 0.006896552 0.006896552 0.008275862 0.001379310 0.001379310 0.001379310
#>
#> $mids
#> [1] 2.5 7.5 12.5 17.5 22.5 27.5 32.5 37.5 42.5 47.5 52.5 57.5
#>
#> $xname
#> [1] "table(cassava_EC[, quant][, i])"
#>
#> $equidist
#> [1] TRUE
#>
#> attr(,"class")
#> [1] "histogram"
#>
#> [[6]]
#> $breaks
#> [1] 0 5 10 15 20 25 30 35 40 45 50 55 60
#>
#> $counts
#> [1] 203 37 18 6 5 3 3 6 0 0 1 1
#>
#> $density
#> [1] 0.1434628975 0.0261484099 0.0127208481 0.0042402827 0.0035335689
#> [6] 0.0021201413 0.0021201413 0.0042402827 0.0000000000 0.0000000000
#> [11] 0.0007067138 0.0007067138
#>
#> $mids
#> [1] 2.5 7.5 12.5 17.5 22.5 27.5 32.5 37.5 42.5 47.5 52.5 57.5
#>
#> $xname
#> [1] "table(cassava_EC[, quant][, i])"
#>
#> $equidist
#> [1] TRUE
#>
#> attr(,"class")
#> [1] "histogram"
#>
#> [[7]]
#> $breaks
#> [1] 0 5 10 15 20 25 30 35
#>
#> $counts
#> [1] 127 32 32 25 8 5 1
#>
#> $density
#> [1] 0.1104347826 0.0278260870 0.0278260870 0.0217391304 0.0069565217
#> [6] 0.0043478261 0.0008695652
#>
#> $mids
#> [1] 2.5 7.5 12.5 17.5 22.5 27.5 32.5
#>
#> $xname
#> [1] "table(cassava_EC[, quant][, i])"
#>
#> $equidist
#> [1] TRUE
#>
#> attr(,"class")
#> [1] "histogram"
#>
#> [[8]]
#> $breaks
#> [1] 0 5 10 15 20 25 30 35
#>
#> $counts
#> [1] 347 41 19 3 7 4 2
#>
#> $density
#> [1] 0.1640661939 0.0193853428 0.0089834515 0.0014184397 0.0033096927
#> [6] 0.0018912530 0.0009456265
#>
#> $mids
#> [1] 2.5 7.5 12.5 17.5 22.5 27.5 32.5
#>
#> $xname
#> [1] "table(cassava_EC[, quant][, i])"
#>
#> $equidist
#> [1] TRUE
#>
#> attr(,"class")
#> [1] "histogram"
#>
#> [[9]]
#> $breaks
#> [1] 0 100 200 300 400 500 600 700
#>
#> $counts
#> [1] 12 2 2 0 0 0 1
#>
#> $density
#> [1] 0.0070588235 0.0011764706 0.0011764706 0.0000000000 0.0000000000
#> [6] 0.0000000000 0.0005882353
#>
#> $mids
#> [1] 50 150 250 350 450 550 650
#>
#> $xname
#> [1] "table(cassava_EC[, quant][, i])"
#>
#> $equidist
#> [1] TRUE
#>
#> attr(,"class")
#> [1] "histogram"
#>
#> [[10]]
#> $breaks
#> [1] 0 5 10 15 20 25 30
#>
#> $counts
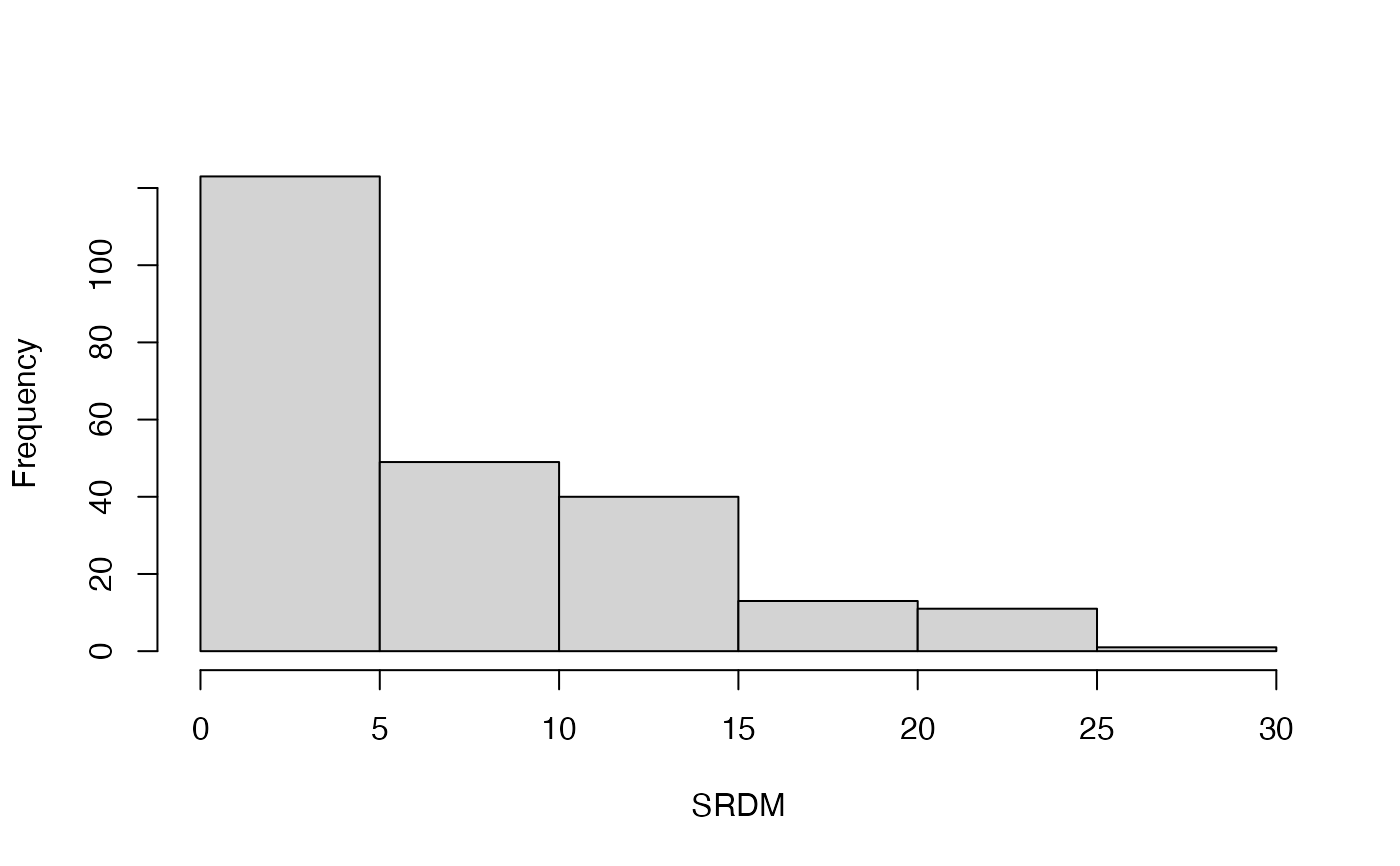
#> [1] 123 49 40 13 11 1
#>
#> $density
#> [1] 0.1037974684 0.0413502110 0.0337552743 0.0109704641 0.0092827004
#> [6] 0.0008438819
#>
#> $mids
#> [1] 2.5 7.5 12.5 17.5 22.5 27.5
#>
#> $xname
#> [1] "table(cassava_EC[, quant][, i])"
#>
#> $equidist
#> [1] TRUE
#>
#> attr(,"class")
#> [1] "histogram"
#>
#> [[1]]
#> $breaks
#> [1] 0 20 40 60 80 100 120
#>
#> $counts
#> [1] 18 5 5 6 7 1
#>
#> $density
#> [1] 0.021428571 0.005952381 0.005952381 0.007142857 0.008333333 0.001190476
#>
#> $mids
#> [1] 10 30 50 70 90 110
#>
#> $xname
#> [1] "table(cassava_EC[, quant][, i])"
#>
#> $equidist
#> [1] TRUE
#>
#> attr(,"class")
#> [1] "histogram"
#>
#> [[2]]
#> $breaks
#> [1] 0 20 40 60 80 100 120 140 160 180
#>
#> $counts
#> [1] 72 9 5 2 0 1 2 0 1
#>
#> $density
#> [1] 0.0391304348 0.0048913043 0.0027173913 0.0010869565 0.0000000000
#> [6] 0.0005434783 0.0010869565 0.0000000000 0.0005434783
#>
#> $mids
#> [1] 10 30 50 70 90 110 130 150 170
#>
#> $xname
#> [1] "table(cassava_EC[, quant][, i])"
#>
#> $equidist
#> [1] TRUE
#>
#> attr(,"class")
#> [1] "histogram"
#>
#> [[3]]
#> $breaks
#> [1] 0 10 20 30 40 50 60 70 80
#>
#> $counts
#> [1] 77 13 8 15 5 1 2 2
#>
#> $density
#> [1] 0.0626016260 0.0105691057 0.0065040650 0.0121951220 0.0040650407
#> [6] 0.0008130081 0.0016260163 0.0016260163
#>
#> $mids
#> [1] 5 15 25 35 45 55 65 75
#>
#> $xname
#> [1] "table(cassava_EC[, quant][, i])"
#>
#> $equidist
#> [1] TRUE
#>
#> attr(,"class")
#> [1] "histogram"
#>
#> [[4]]
#> $breaks
#> [1] 0 10 20 30 40 50 60 70 80 90 100
#>
#> $counts
#> [1] 204 24 7 2 2 2 2 0 0 1
#>
#> $density
#> [1] 0.0836065574 0.0098360656 0.0028688525 0.0008196721 0.0008196721
#> [6] 0.0008196721 0.0008196721 0.0000000000 0.0000000000 0.0004098361
#>
#> $mids
#> [1] 5 15 25 35 45 55 65 75 85 95
#>
#> $xname
#> [1] "table(cassava_EC[, quant][, i])"
#>
#> $equidist
#> [1] TRUE
#>
#> attr(,"class")
#> [1] "histogram"
#>
#> [[5]]
#> $breaks
#> [1] 0 5 10 15 20 25 30 35 40 45 50 55 60
#>
#> $counts
#> [1] 75 18 11 9 9 4 5 5 6 1 1 1
#>
#> $density
#> [1] 0.103448276 0.024827586 0.015172414 0.012413793 0.012413793 0.005517241
#> [7] 0.006896552 0.006896552 0.008275862 0.001379310 0.001379310 0.001379310
#>
#> $mids
#> [1] 2.5 7.5 12.5 17.5 22.5 27.5 32.5 37.5 42.5 47.5 52.5 57.5
#>
#> $xname
#> [1] "table(cassava_EC[, quant][, i])"
#>
#> $equidist
#> [1] TRUE
#>
#> attr(,"class")
#> [1] "histogram"
#>
#> [[6]]
#> $breaks
#> [1] 0 5 10 15 20 25 30 35 40 45 50 55 60
#>
#> $counts
#> [1] 203 37 18 6 5 3 3 6 0 0 1 1
#>
#> $density
#> [1] 0.1434628975 0.0261484099 0.0127208481 0.0042402827 0.0035335689
#> [6] 0.0021201413 0.0021201413 0.0042402827 0.0000000000 0.0000000000
#> [11] 0.0007067138 0.0007067138
#>
#> $mids
#> [1] 2.5 7.5 12.5 17.5 22.5 27.5 32.5 37.5 42.5 47.5 52.5 57.5
#>
#> $xname
#> [1] "table(cassava_EC[, quant][, i])"
#>
#> $equidist
#> [1] TRUE
#>
#> attr(,"class")
#> [1] "histogram"
#>
#> [[7]]
#> $breaks
#> [1] 0 5 10 15 20 25 30 35
#>
#> $counts
#> [1] 127 32 32 25 8 5 1
#>
#> $density
#> [1] 0.1104347826 0.0278260870 0.0278260870 0.0217391304 0.0069565217
#> [6] 0.0043478261 0.0008695652
#>
#> $mids
#> [1] 2.5 7.5 12.5 17.5 22.5 27.5 32.5
#>
#> $xname
#> [1] "table(cassava_EC[, quant][, i])"
#>
#> $equidist
#> [1] TRUE
#>
#> attr(,"class")
#> [1] "histogram"
#>
#> [[8]]
#> $breaks
#> [1] 0 5 10 15 20 25 30 35
#>
#> $counts
#> [1] 347 41 19 3 7 4 2
#>
#> $density
#> [1] 0.1640661939 0.0193853428 0.0089834515 0.0014184397 0.0033096927
#> [6] 0.0018912530 0.0009456265
#>
#> $mids
#> [1] 2.5 7.5 12.5 17.5 22.5 27.5 32.5
#>
#> $xname
#> [1] "table(cassava_EC[, quant][, i])"
#>
#> $equidist
#> [1] TRUE
#>
#> attr(,"class")
#> [1] "histogram"
#>
#> [[9]]
#> $breaks
#> [1] 0 100 200 300 400 500 600 700
#>
#> $counts
#> [1] 12 2 2 0 0 0 1
#>
#> $density
#> [1] 0.0070588235 0.0011764706 0.0011764706 0.0000000000 0.0000000000
#> [6] 0.0000000000 0.0005882353
#>
#> $mids
#> [1] 50 150 250 350 450 550 650
#>
#> $xname
#> [1] "table(cassava_EC[, quant][, i])"
#>
#> $equidist
#> [1] TRUE
#>
#> attr(,"class")
#> [1] "histogram"
#>
#> [[10]]
#> $breaks
#> [1] 0 5 10 15 20 25 30
#>
#> $counts
#> [1] 123 49 40 13 11 1
#>
#> $density
#> [1] 0.1037974684 0.0413502110 0.0337552743 0.0109704641 0.0092827004
#> [6] 0.0008438819
#>
#> $mids
#> [1] 2.5 7.5 12.5 17.5 22.5 27.5
#>
#> $xname
#> [1] "table(cassava_EC[, quant][, i])"
#>
#> $equidist
#> [1] TRUE
#>
#> attr(,"class")
#> [1] "histogram"
#>