Compute Principal Component Analysis Statistics (Mardia et al. 1979) to compare the probability distributions of quantitative traits between entire collection (EC) and core set (CS).
Usage
pca.evaluate.core(
data,
names,
quantitative,
selected,
center = TRUE,
scale = TRUE,
npc.plot = 6
)Arguments
- data
The data as a data frame object. The data frame should possess one row per individual and columns with the individual names and multiple trait/character data.
- names
Name of column with the individual names as a character string.
- quantitative
Name of columns with the quantitative traits as a character vector.
- selected
Character vector with the names of individuals selected in core collection and present in the
namescolumn.- center
either a logical value or numeric-alike vector of length equal to the number of columns of
x, where ‘numeric-alike’ means thatas.numeric(.)will be applied successfully ifis.numeric(.)is not true.- scale
either a logical value or a numeric-alike vector of length equal to the number of columns of
x.- npc.plot
The number of principal components for which eigen values are to be plotted. The default value is 6.
Value
A list with the following components.
- EC PC Importance
A data frame of importance of principal components for EC
- EC PC Loadings
A data frame with eigen vectors of principal components for EC
- CS PC Importance
A data frame of importance of principal components for CS
- CS PC Loadings
A data frame with eigen vectors of principal components for CS
- Scree Plot
The scree plot of principal components for EC and CS as a
ggplotobject.- PC Loadings Plot
A plot of the eigen vector values of principal components for EC and CS as specified by
npc.plotas aggplot2object.
Note
If there are missing values in the quantitative data, then they will be
imputed using the imputePCA function from
missMDA.
References
Mardia KV, Kent JT, Bibby JM (1979). Multivariate analysis. Academic Press, London; New York. ISBN 0-12-471250-9 978-0-12-471250-8 0-12-471252-5 978-0-12-471252-2.
Examples
data("cassava_CC")
data("cassava_EC")
ec <- cbind(genotypes = rownames(cassava_EC), cassava_EC)
ec$genotypes <- as.character(ec$genotypes)
rownames(ec) <- NULL
core <- rownames(cassava_CC)
quant <- c("NMSR", "TTRN", "TFWSR", "TTRW", "TFWSS", "TTSW", "TTPW", "AVPW",
"ARSR", "SRDM")
qual <- c("CUAL", "LNGS", "PTLC", "DSTA", "LFRT", "LBTEF", "CBTR", "NMLB",
"ANGB", "CUAL9M", "LVC9M", "TNPR9M", "PL9M", "STRP", "STRC",
"PSTR")
ec[, qual] <- lapply(ec[, qual],
function(x) factor(as.factor(x)))
pca.evaluate.core(data = ec, names = "genotypes",
quantitative = quant, selected = core,
center = TRUE, scale = TRUE, npc.plot = 4)
#> $`EC PC Importance`
#> PC1 PC2 PC3 PC4 PC5 PC6
#> Standard deviation 2.303455 1.237223 0.996322 0.9455592 0.8817511 0.6251589
#> Proportion of Variance 0.530590 0.153070 0.099270 0.0894100 0.0777500 0.0390800
#> Cumulative Proportion 0.530590 0.683660 0.782930 0.8723400 0.9500900 0.9891700
#> PC7 PC8 PC9 PC10
#> Standard deviation 0.271212 0.1864651 3.833103e-15 7.684284e-16
#> Proportion of Variance 0.007360 0.0034800 0.000000e+00 0.000000e+00
#> Cumulative Proportion 0.996520 1.0000000 1.000000e+00 1.000000e+00
#>
#> $`EC PC Loadings`
#> PC1 PC2 PC3 PC4 PC5 PC6
#> NMSR 0.276022186 0.55062492 0.01668655 -0.1871023 0.248244608 -0.21686806
#> TTRN 0.293942565 0.20622412 0.17262243 0.3680853 0.581178024 -0.33878178
#> TFWSR 0.371192088 0.06599302 -0.07379106 -0.1852754 -0.463167012 -0.33058227
#> TTRW 0.334894681 -0.33211659 0.03783194 0.3153475 -0.297171591 -0.40373685
#> TFWSS 0.385024604 0.13341651 -0.08631304 -0.3426523 0.050559491 0.39246657
#> TTSW 0.362255093 -0.27969168 0.02022637 0.1585548 0.250560331 0.51748203
#> TTPW 0.403699747 0.11088360 -0.08613665 -0.2920918 -0.184053823 0.08318831
#> AVPW 0.380627616 -0.33011132 0.03067827 0.2497406 0.002748448 0.10913947
#> ARSR 0.011265368 0.52673026 0.37858111 0.5081543 -0.444763915 0.34913640
#> SRDM -0.004586239 0.21186429 -0.89638345 0.3837654 -0.027148446 0.05797814
#> PC7 PC8 PC9 PC10
#> NMSR -0.66616815 -0.181386616 3.702136e-16 -2.125739e-16
#> TTRN 0.49417918 0.095026370 -3.246553e-16 4.790702e-16
#> TFWSR 0.32960316 -0.499641762 3.415602e-01 1.390591e-01
#> TTRW -0.31608453 0.416500300 1.469979e-01 -3.610597e-01
#> TFWSS 0.10285083 0.558642359 4.440363e-01 1.807801e-01
#> TTSW -0.07640644 -0.454657280 1.776166e-01 -4.362661e-01
#> TTPW 0.21455970 0.104941479 -7.375515e-01 -3.002787e-01
#> AVPW -0.20120313 -0.065453705 -2.983792e-01 7.328858e-01
#> ARSR 0.01979215 0.027945977 3.446846e-17 4.351411e-17
#> SRDM 0.01152032 0.009060183 -1.901142e-16 -6.838485e-17
#>
#> $`CS PC Importance`
#> PC1 PC2 PC3 PC4 PC5 PC6
#> Standard deviation 2.280951 1.312257 1.000457 0.8959377 0.8451336 0.6538375
#> Proportion of Variance 0.520270 0.172200 0.100090 0.0802700 0.0714300 0.0427500
#> Cumulative Proportion 0.520270 0.692480 0.792570 0.8728400 0.9442600 0.9870100
#> PC7 PC8 PC9 PC10
#> Standard deviation 0.3207286 0.1643265 9.704593e-16 2.283229e-16
#> Proportion of Variance 0.0102900 0.0027000 0.000000e+00 0.000000e+00
#> Cumulative Proportion 0.9973000 1.0000000 1.000000e+00 1.000000e+00
#>
#> $`CS PC Loadings`
#> PC1 PC2 PC3 PC4 PC5 PC6
#> NMSR 0.23686459 0.57588211 -0.118000407 0.18406534 0.22404134 -0.154356246
#> TTRN 0.27243380 0.28489364 -0.238654425 -0.39636509 0.59296937 -0.185643439
#> TFWSR 0.38732336 0.05088131 0.173676592 0.15588284 -0.33283776 -0.410239780
#> TTRW 0.34195710 -0.29955595 0.132403764 -0.37824392 -0.14609160 -0.425032356
#> TFWSS 0.38798499 0.11032791 0.002061297 0.40943370 -0.02293943 0.339541418
#> TTSW 0.36275510 -0.26716740 -0.043442847 -0.11254725 0.18634678 0.589974192
#> TTPW 0.41017366 0.08800885 0.085090829 0.31069206 -0.17398680 -0.002991297
#> AVPW 0.38441017 -0.30812430 0.044838607 -0.26192922 0.02880417 0.110848682
#> ARSR 0.02642625 0.53641424 0.049592980 -0.54225171 -0.54787309 0.334180984
#> SRDM -0.06233279 0.14455341 0.931570561 -0.04658879 0.31551819 0.073690249
#> PC7 PC8 PC9 PC10
#> NMSR 0.69455927 0.089542525 0.000000e+00 0.000000e+00
#> TTRN -0.49373454 -0.026538022 3.957456e-16 2.284470e-16
#> TFWSR -0.24125034 0.553541017 5.069141e-02 3.829779e-01
#> TTRW 0.26321979 -0.434887362 -4.115317e-01 5.447083e-02
#> TFWSS -0.17717813 -0.557775840 6.032185e-02 4.557367e-01
#> TTSW 0.13648832 0.424831173 -4.439792e-01 5.876562e-02
#> TTPW -0.21841109 -0.053240284 -1.049261e-01 -7.927258e-01
#> AVPW 0.21517212 0.012287406 7.850518e-01 -1.039104e-01
#> ARSR -0.04644065 -0.032656495 1.513333e-16 1.255519e-16
#> SRDM -0.01212024 -0.009789162 2.132957e-16 4.099931e-17
#>
#> $`Scree Plot`
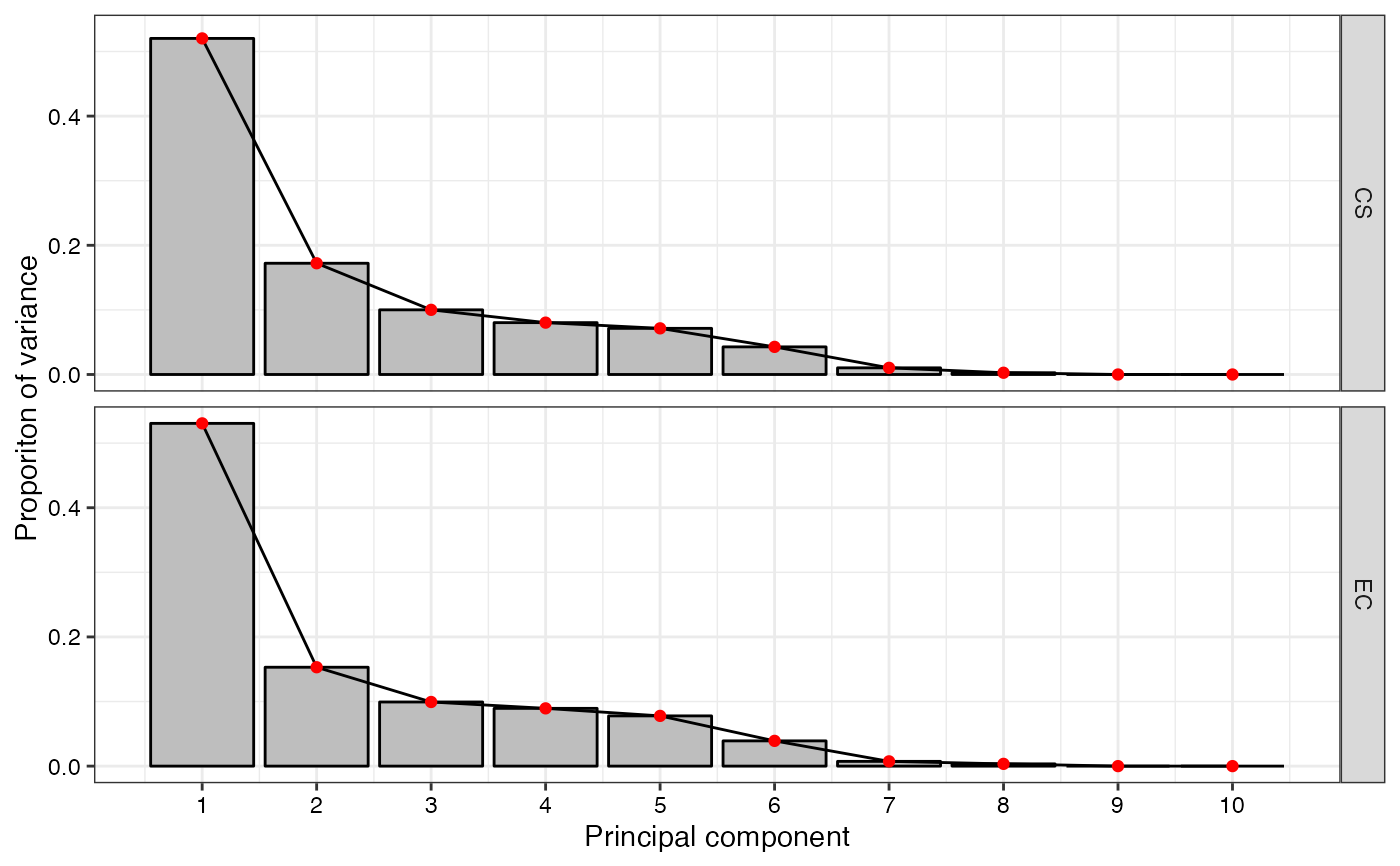 #>
#> $`PC Loadings Plot`
#>
#> $`PC Loadings Plot`
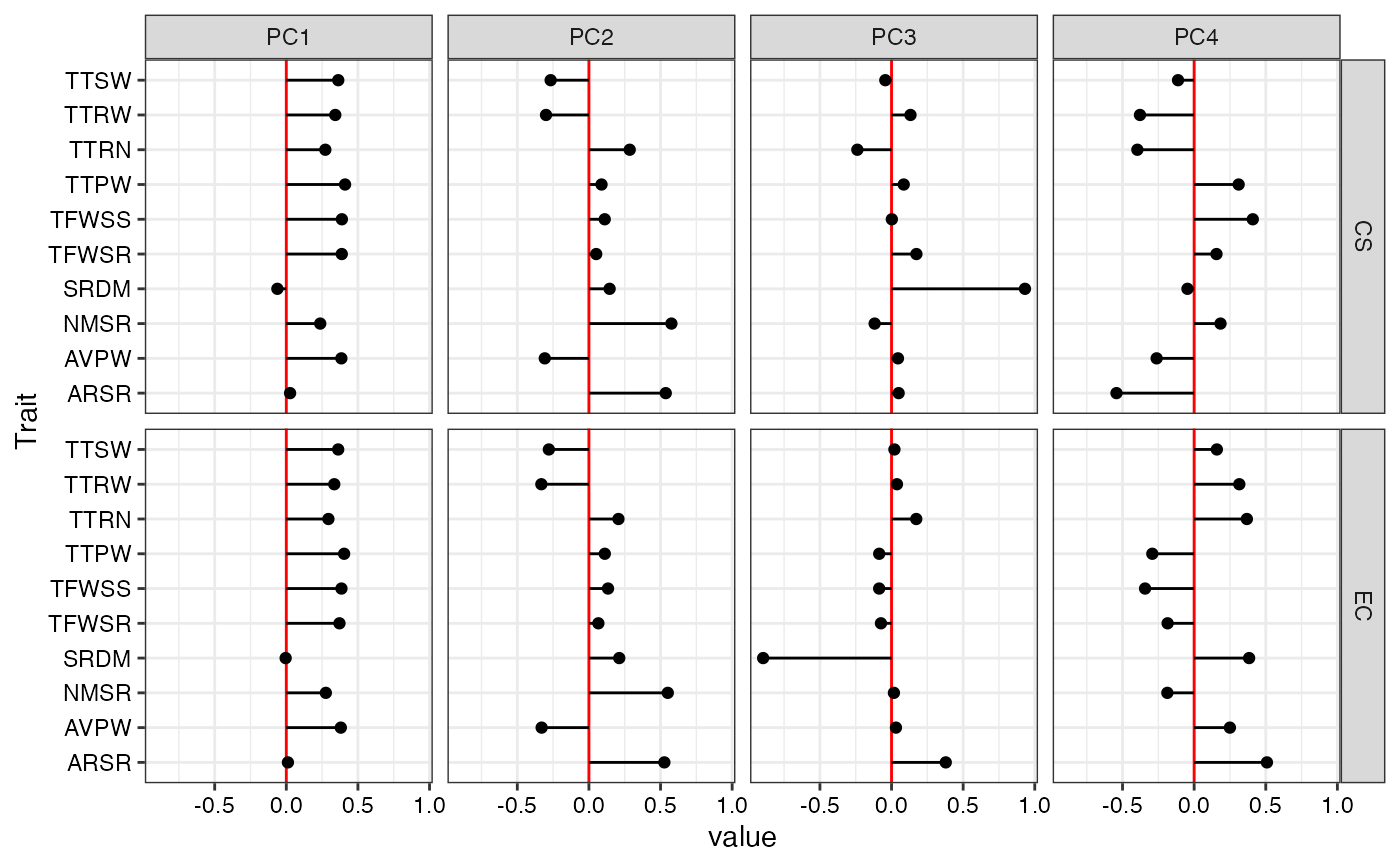 #>
#>